Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 Å Resolution
- PMID: 32004465
- PMCID: PMC7166091
- DOI: 10.1016/j.str.2019.12.002
Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 Å Resolution
Abstract
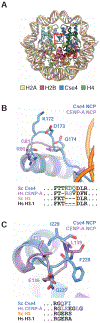
Kinetochores mediate chromosome segregation during cell division. They assemble on centromeric nucleosomes and capture spindle microtubules. In budding yeast, a kinetochore links a single nucleosome, containing the histone variant Cse4CENP-A instead of H3, with a single microtubule. Conservation of most kinetochore components from yeast to metazoans suggests that the yeast kinetochore represents a module of the more complex metazoan arrangements. We describe here a streamlined protocol for reconstituting a yeast centromeric nucleosome and a systematic exploration of cryo-grid preparation. These developments allowed us to obtain a high-resolution cryoelectron microscopy reconstruction. As suggested by previous work, fewer base pairs are in tight association with the histone octamer than there are in canonical nucleosomes. Weak binding of the end DNA sequences may contribute to specific recognition by other inner kinetochore components. The centromeric nucleosome structure and the strategies we describe will facilitate studies of many other aspects of kinetochore assembly and chromatin biochemistry.
Keywords: CENP-A; Cse4; NCP; cryo-EM; histones; kinetochore; nucleosome; structure.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
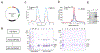


References
-
- Anderson M, Huh JH, Ngo T, Lee A, Hernandez G, Pang J, Perkins J, and Dutnall RN (2010). Co-expression as a convenient method for the production and purification of core histones in bacteria. Protein Expression and Purification 72, 194–204. - PubMed
-
- Black BE, foltz DR, Chakravarthy S, Luger K, woods VL, and Cleveland DW (2014). Structural determinants for generating centromeric chromatin. 430, 1–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

