Prediction of blood-based biomarkers and subsequent design of bisulfite PCR-LDR-qPCR assay for breast cancer detection
- PMID: 32005108
- PMCID: PMC6995062
- DOI: 10.1186/s12885-020-6574-4
Prediction of blood-based biomarkers and subsequent design of bisulfite PCR-LDR-qPCR assay for breast cancer detection
Abstract
Background: Interrogation of site-specific CpG methylation in circulating tumor DNAs (ctDNAs) has been employed in a number of studies for early detection of breast cancer (BrCa). In many of these studies, the markers were identified based on known biology of BrCa progression, and interrogated using methyl-specific PCR (MSP), a technique involving bisulfite conversion, PCR, and qPCR.
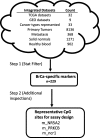
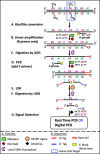
Methods: In this report, we are demonstrating the development of a novel assay (Multiplex Bisulfite PCR-LDR-qPCR) which can potentially offer improvements to MSP, by integrating additional steps such as ligase detection reaction (LDR), methylated CpG target enrichment, carryover protection (use of uracil DNA glycosylase), and minimization of primer-dimer formation (use of ribose primers and RNAseH2). The assay is designed to for breast cancer-specific CpG markers identified through integrated analyses of publicly available genome-wide methylation datasets for 31 types of primary tumors (including BrCa), as well as matching normal tissues, and peripheral blood.
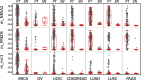
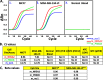
Results: Our results indicate that the PCR-LDR-qPCR assay is capable of detecting ~ 30 methylated copies of each of 3 BrCa-specific CpG markers, when mixed with excess amount unmethylated CpG markers (~ 3000 copies each), which is a reasonable approximation of BrCa ctDNA overwhelmed with peripheral blood cell-free DNA (cfDNA) when isolated from patient plasma. The bioinformatically-identified CpG markers are located in promoter regions of NR5A2 and PRKCB, and a non-coding region of chromosome 1 (upstream of EFNA3). Additional bioinformatic analyses would reveal that these methylation markers are independent of patient race and age, and positively associated with signaling pathways associated with BrCa progression (such as those related to retinoid nuclear receptor, PTEN, p53, pRB, and p27).
Conclusion: This report demonstrates the potential utilization of bisulfite PCR-LDR-qPCR assay, along with bioinformatically-driven biomarker discovery, in blood-based BrCa detection.
Keywords: Biomarker; Breast cancer; Early detection; Ligase detection reaction; Methylation.
Conflict of interest statement
MDB, AHM, JH, PBF, SFG and FB are shareholders in AcuamarkDx.
Figures


References
-
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72(5):1117–1130. doi: 10.1086/375033. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

