Basal hsp70 expression levels do not explain adaptive variation of the warm- and cold-climate O3 + 4 + 7 and OST gene arrangements of Drosophila subobscura
- PMID: 32005133
- PMCID: PMC6995229
- DOI: 10.1186/s12862-020-1584-z
Basal hsp70 expression levels do not explain adaptive variation of the warm- and cold-climate O3 + 4 + 7 and OST gene arrangements of Drosophila subobscura
Abstract
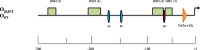
Background: Drosophila subobscura exhibits a rich inversion polymorphism, with some adaptive inversions showing repeatable spatiotemporal patterns in frequencies related to temperature. Previous studies reported increased basal HSP70 protein levels in homokaryotypic strains for a warm-climate arrangement compared to a cold-climate one. These findings do not match the similar hsp70 genomic organization between arrangements, where gene expression levels are expected to be similar. In order to test this hypothesis and understand the molecular basis for hsp70 expression, we compared basal hsp70 mRNA levels in males and females, and analysed the 5' and 3' regulatory regions of hsp70 genes in warm- and cold-climate isochromosomal O3 + 4 + 7 and OST lines of D. subobscura.
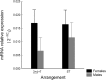
Results: We observed comparable mRNA levels between the two arrangements and a sex-biased hsp70 gene expression. The number of heat-shock elements (HSEs) and GAGA sites on the promoters were identical amongst the OST and O3 + 4 + 7 lines analysed. This is also true for 3' AU-rich elements where most A and B copies of hsp70 have, respectively, two and one element in both arrangements. Beyond the regulatory elements, the only notable difference between both arrangements is the presence in 3' UTR of a 14 bp additional fragment after the stop codon in the hsp70A copy in five O3 + 4 + 7 lines, which was not found in any of the six OST lines.
Conclusions: The equivalent hsp70 mRNA amounts in OST and O3 + 4 + 7 arrangements provide the first evidence of a parallelism between gene expression and genetic organization in D. subobscura lines having these arrangements. This is reinforced by the lack of important differential features in the number and structure of regulatory elements between both arrangements, despite the genetic differentiation observed when the complete 5' and 3' regulatory regions were considered. Therefore, the basal levels of hsp70 mRNA cannot account, in principle, for the adaptive variation of the two arrangements studied. Consequently, further studies are necessary to understand the intricate molecular mechanisms of hsp70 gene regulation in D. subobscura.
Keywords: Chromosomal arrangements; Drosophila subobscura; Thermal adaptation; hsp70 expression.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Menozzi P, Krimbas CB. The inversion polymorphism of D. subobscura revisited: Synthetic maps of gene arrangement frequencies and their interpretation. J Evol Biol. 1992;5:625–641. doi: 10.1046/j.1420-9101.1992.5040625.x. - DOI
-
- . Powell JR: Progress and Prospects in Evolutionary Biology. The Drosophila Model, New York: Oxford Univ Press; 1997.
-
- Krimbas CB. Drosophila subobscura, Biology, Genetics and Inversion Polymorphism. Hamburg: Verlag Dr. Kovač; 1993.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

