A systemic approach to screening high-throughput RT-qPCR data for a suitable set of reference circulating miRNAs
- PMID: 32005151
- PMCID: PMC6995162
- DOI: 10.1186/s12864-020-6530-3
A systemic approach to screening high-throughput RT-qPCR data for a suitable set of reference circulating miRNAs
Abstract
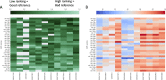
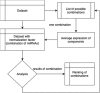
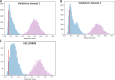
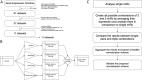
Background: The consensus on how to choose a reference gene for serum or plasma miRNA expression qPCR studies has not been reached and none of the potential candidates have yet been convincingly validated. We proposed a new in silico approach of finding a suitable reference for human, circulating miRNAs and identified a new set of endogenous reference miRNA based on miRNA profiling experiments from Gene Expression Omnibus. We used 3 known normalization algorithms (NormFinder, BestKeeper, GeNorm) to calculate a new normalization score. We searched for a universal set of endogenous miRNAs and validated our findings on 2 new datasets using our approach.
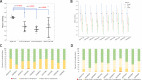
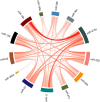
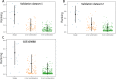
Results: We discovered and validated a set of 13 miRNAs (miR-222, miR-92a, miR-27a, miR-17, miR-24, miR-320a, miR-25, miR-126, miR-19b, miR-199a-3p, miR-30b, miR-30c, miR-374a) that can be used to create a reliable reference combination of 3 miRNAs. We showed that on average the mean of 3 miRNAs (p = 0.0002) and 2 miRNAs (p = 0.0031) were a better reference than single miRNA. The arithmetic means of 3 miRNAs: miR-24, miR-222 and miR-27a was shown to be the most stable combination of 3 miRNAs in validation sets.
Conclusions: No single miRNA was suitable as a universal reference in serum miRNA qPCR profiling, but it was possible to designate a set of miRNAs, which consistently contributed to most stable combinations.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of tranformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79(2):137–158. doi: 10.1084/jem.79.2.137. - DOI - PMC - PubMed
-
- Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli. J Biol Chem. 1958;233(1):163–170. - PubMed
-
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11(9):1026–1030. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

