Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis
- PMID: 32005165
- PMCID: PMC6993422
- DOI: 10.1186/s12870-020-2252-6
Characterization of wheat homeodomain-leucine zipper family genes and functional analysis of TaHDZ5-6A in drought tolerance in transgenic Arabidopsis
Abstract
Background: Many studies in Arabidopsis and rice have demonstrated that HD-Zip transcription factors play important roles in plant development and responses to abiotic stresses. Although common wheat (Triticum aestivum L.) is one of the most widely cultivated and consumed food crops in the world, the function of the HD-Zip proteins in wheat is still largely unknown.
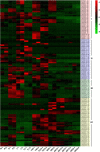
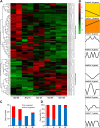
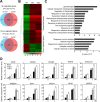
Results: To explore the potential biological functions of HD-Zip genes in wheat, we performed a bioinformatics and gene expression analysis of the HD-Zip family. We identified 113 HD-Zip members from wheat and classified them into four subfamilies (I-IV) based on phylogenic analysis against proteins from Arabidopsis, rice, and maize. Most HD-Zip genes are represented by two to three homeoalleles in wheat, which are named as TaHDZX_ZA, TaHDZX_ZB, or TaHDZX_ZD, where X denotes the gene number and Z the wheat chromosome on which it is located. TaHDZs in the same subfamily have similar protein motifs and intron/exon structures. The expression profiles of TaHDZ genes were analysed in different tissues, at different stages of vegetative growth, during seed development, and under drought stress. We found that most TaHDZ genes, especially those in subfamilies I and II, were induced by drought stress, suggesting the potential importance of subfamily I and II TaHDZ members in the responses to abiotic stress. Compared with wild-type (WT) plants, transgenic Arabidopsis plants overexpressing TaHDZ5-6A displayed enhanced drought tolerance, lower water loss rates, higher survival rates, and higher proline content under drought conditions. Additionally, the transcriptome analysis identified a number of differentially expressed genes between 35S::TaHDZ5-6A transgenic and wild-type plants, many of which are involved in stress response.
Conclusions: Our results will facilitate further functional analysis of wheat HD-Zip genes, and also indicate that TaHDZ5-6A may participate in regulating the plant response to drought stress. Our experiments show that TaHDZ5-6A holds great potential for genetic improvement of abiotic stress tolerance in crops.
Keywords: Drought tolerance; Expression profiles; HD-zip gene family; Phylogenetic relationships; TaHDZ5-6A; Wheat.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

