Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor)
- PMID: 32005168
- PMCID: PMC6995107
- DOI: 10.1186/s12864-020-6538-8
Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor)
Abstract
Background: In sorghum (Sorghum bicolor), one paramount breeding objective is to increase grain quality. The nutritional quality and end use value of sorghum grains are primarily influenced by the proportions of tannins, starch and proteins, but the genetic basis of these grain quality traits remains largely unknown. This study aimed to dissect the natural variation of sorghum grain quality traits and identify the underpinning genetic loci by genome-wide association study.
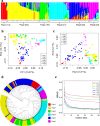
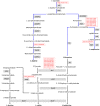
Results: Levels of starch, tannins and 17 amino acids were quantified in 196 diverse sorghum inbred lines, and 44 traits based on known metabolic pathways and biochemical interactions amongst the 17 amino acids calculated. A Genome-wide association study (GWAS) with 3,512,517 SNPs from re-sequencing data identified 14, 15 and 711 significant SNPs which represented 14, 14, 492 genetic loci associated with levels of tannins, starch and amino acids in sorghum grains, respectively. Amongst these significant SNPs, two SNPs were associated with tannin content on chromosome 4 and colocalized with three previously identified loci for Tannin1, and orthologs of Zm1 and TT16 genes. One SNP associated with starch content colocalized with sucrose phosphate synthase gene. Furthermore, homologues of opaque1 and opaque2 genes associated with amino acid content were identified. Using the KEGG pathway database, six and three candidate genes of tannins and starch were mapped into 12 and 3 metabolism pathways, respectively. Thirty-four candidate genes were mapped into 16 biosynthetic and catabolic pathways of amino acids. We finally reconstructed the biosynthetic pathways for aspartate and branched-chain amino acids based on 15 candidate genes identified in this study.
Conclusion: Promising candidate genes associated with grain quality traits have been identified in the present study. Some of them colocalized with previously identified genetic regions, but novel candidate genes involved in various metabolic pathways which influence grain quality traits have been dissected. Our study acts as an entry point for further validation studies to elucidate the complex mechanisms controlling grain quality traits such as tannins, starch and amino acids in sorghum.
Keywords: Amino acids; Genome-wide association study; Grain quality; Sorghum; Starch; Tannins.
Conflict of interest statement
The authors declare they have no competing interests.
Figures




References
-
- Sukumaran S, Xiang W, Bean SR, Pedersen JF, Kresovich S, Tuinstra MR, et al. Association mapping for grain quality in a diverse Sorghum collection. Plant Genome. 2012;5:126–135. doi: 10.3835/plantgenome2012.07.0016. - DOI
MeSH terms
Substances
Grants and funding
- 2018YFD1000701, 2018YFD1000700/National Key R&D Program of China
- 2015BAD15B03/Ministry of Science and Technology of the People's Republic of China
- KFJ-FP-201809; KFJ-STS-ZDTP-056/Science and Technology Service Network Initiative Project of Chinese Academy of Sciences
- 31461143023/National Natural Science Foundation of China
- SAJC201603/Sino-Africa Joint Research Center, Chinese Academy of Sciences
LinkOut - more resources
Full Text Sources

