In vivo model to study the impact of genetic variation on clinical outcome of mastitis in uniparous dairy cows
- PMID: 32005239
- PMCID: PMC6995066
- DOI: 10.1186/s12917-020-2251-8
In vivo model to study the impact of genetic variation on clinical outcome of mastitis in uniparous dairy cows
Abstract
Background: In dairy herds, mastitis causes detrimental economic losses. Genetic selection offers a sustainable tool to select animals with reduced susceptibility towards postpartum diseases. Studying underlying mechanisms is important to assess the physiological processes that cause differences between selected haplotypes. Therefore, the objective of this study was to establish an in vivo infection model to study the impact of selecting for alternative paternal haplotypes in a particular genomic region on cattle chromosome 18 for mastitis susceptibility under defined conditions in uniparous dairy cows.
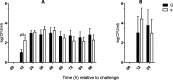
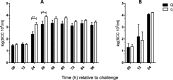
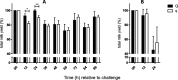
Results: At the start of pathogen challenge, no significant differences between the favorable (Q) and unfavorable (q) haplotypes were detected. Intramammary infection (IMI) with Staphylococcus aureus 1027 (S. aureus, n = 24, 96 h) or Escherichia coli 1303 (E. coli, n = 12, 24 h) was successfully induced in all uniparous cows. This finding was confirmed by clinical signs of mastitis and repeated recovery of the respective pathogen from milk samples of challenged quarters in each animal. After S. aureus challenge, Q-uniparous cows showed lower somatic cell counts 24 h and 36 h after challenge (P < 0.05), lower bacterial shedding in milk 12 h after challenge (P < 0.01) and a minor decrease in total milk yield 12 h and 24 h after challenge (P < 0.01) compared to q-uniparous cows.
Conclusion: An in vivo infection model to study the impact of genetic selection for mastitis susceptibility under defined conditions in uniparous dairy cows was successfully established and revealed significant differences between the two genetically selected haplotype groups. This result might explain their differences in susceptibility towards IMI. These clinical findings form the basis for further in-depth molecular analysis to clarify the underlying genetic mechanisms for mastitis resistance.
Keywords: BTA18; Escherichia coli; Genetic selection; Haplotype; Intramammary infection model; Mastitis; Somatic cell count; Staphylococcus aureus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

