Amphotericin B biosynthesis in Streptomyces nodosus: quantitative analysis of metabolism via LC-MS/MS based metabolomics for rational design
- PMID: 32005241
- PMCID: PMC6995120
- DOI: 10.1186/s12934-020-1290-y
Amphotericin B biosynthesis in Streptomyces nodosus: quantitative analysis of metabolism via LC-MS/MS based metabolomics for rational design
Abstract
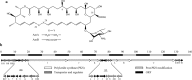
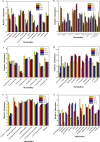
Background: Amphotericin B (AmB) is widely used against fungal infection and produced mainly by Streptomyces nodosus. Various intracellular metabolites of S. nodosus were identified during AmB fermentation, and the key compounds that related to the cell growth and biosynthesis of AmB were analyzed by principal component analysis (PCA) and partial least squares (PLS).
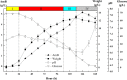
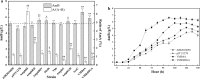
Results: Rational design that based on the results of metabolomics was employed to improve the AmB productivity of Streptomyces nodosus, including the overexpression of genes involved in oxygen-taking, precursor-acquiring and product-exporting. The AmB yield of modified strain S. nodosus VMR4A was 6.58 g/L, which was increased significantly in comparison with that of strain S. nodosus ZJB2016050 (5.16 g/L). This was the highest yield of AmB reported so far, and meanwhile, the amount of by-product amphotericin A (AmA) was decreased by 45%. Moreover, the fermentation time of strain S. nodosus VMR4A was shortened by 24 h compared with that of strain. The results indicated that strain S. nodosus VMR4A was an excellent candidate for the industrial production of AmB because of its high production yield, low by-product content and the fast cell growth.
Conclusions: This study would lay the foundation for improving the AmB productivity through metabolomics analysis and overexpression of key enzymes.
Keywords: Amphotericin B; Metabolomics; Overexpression; Streptomyces nodosus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

