The prognostic value of a seven-lncRNA signature in patients with esophageal squamous cell carcinoma: a lncRNA expression analysis
- PMID: 32005248
- PMCID: PMC6995134
- DOI: 10.1186/s12967-020-02224-z
The prognostic value of a seven-lncRNA signature in patients with esophageal squamous cell carcinoma: a lncRNA expression analysis
Abstract
Background: Long non-coding RNAs (lncRNAs) have been reported to be prognostic biomarkers in many types of cancer. We aimed to identify a lncRNA signature that can predict the prognosis in patients with esophageal squamous cell carcinoma (ESCC).
Methods: Using a custom microarray, we retrospectively analyzed lncRNA expression profiles in 141 samples of ESCC and 81 paired non-cancer specimens from Sun Yat-Sen University Cancer Center (Guangzhou, China), which were used as a training cohort to identify a signature associated with clinical outcomes. Then we conducted quantitative RT-PCR in another 103 samples of ESCC from the same cancer center as an independent cohort to verify the signature.
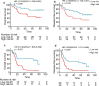
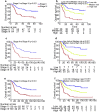
Results: Microarray analysis showed that there were 338 lncRNAs significantly differentially expressed between ESCC and non-cancer esophagus tissues in the training cohort. From these differentially expressed lncRNAs, we found 16 lncRNAs associated with overall survival (OS) of ESCC patients using Cox regression analysis. Then a 7-lncRNA signature for predicting survival was identified from the 16 lncRNAs, which classified ESCC patients into high-risk and low-risk groups. Patients with high-risk have shorter OS (HR: 3.555, 95% CI 2.195-5.757, p < 0.001) and disease-free survival (DFS) (HR: 2.537, 95% CI 1.646-3.909, p < 0.001) when compared with patients with low-risk in the training cohort. In the independent cohort, the 7 lncRNAs were detected by qRT-PCR and used to compute risk score for the patients. The result indicates that patients with high risk also have significantly worse OS (HR = 2.662, 95% CI 1.588-4.464, p < 0.001) and DFS (HR 2.389, 95% CI 1.447-3.946, p < 0.001). The univariate and multivariate Cox regression analyses indicate that the signature is an independent factor for predicting survival of patients with ESCC. Combination of the signature and TNM staging was more powerful in predicting OS than TNM staging alone in both the training (AUC: 0.772 vs 0.681, p = 0.002) and independent cohorts (AUC: 0.772 vs 0.660, p = 0.003).
Conclusions: The 7-lncRNA signature is a potential prognostic biomarker in patients with ESCC and may help in treatment decision when combined with the TNM staging system.
Keywords: Esophageal cancer; Expression profile; Survival; TNM stage; lncRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Ishiguro S, Sasazuki S, Inoue M, Kurahashi N, Iwasaki M, Tsugane S, Group JS. Effect of alcohol consumption, cigarette smoking and flushing response on esophageal cancer risk: a population-based cohort study (JPHC study) Cancer Lett. 2009;275:240–246. doi: 10.1016/j.canlet.2008.10.020. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

