Prognostic DNA methylation markers for hormone receptor breast cancer: a systematic review
- PMID: 32005275
- PMCID: PMC6993426
- DOI: 10.1186/s13058-020-1250-9
Prognostic DNA methylation markers for hormone receptor breast cancer: a systematic review
Abstract
Background: In patients with hormone receptor-positive breast cancer, differentiating between patients with a low and a high risk of recurrence is an ongoing challenge. In current practice, prognostic clinical parameters are used for risk prediction. DNA methylation markers have been proven to be of additional prognostic value in several cancer types. Numerous prognostic DNA methylation markers for breast cancer have been published in the literature. However, to date, none of these markers are used in clinical practice.
Methods: We conducted a systematic review of PubMed and EMBASE to assess the number and level of evidence of published DNA methylation markers for hormone receptor-positive breast cancer. To obtain an overview of the reporting quality of the included studies, all were scored according to the REMARK criteria that were established as reporting guidelines for prognostic biomarker studies.
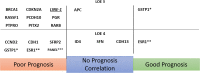
Results: A total of 74 studies were identified reporting on 87 different DNA methylation markers. Assessment of the REMARK criteria showed variation in reporting quality of the studies. Eighteen single markers and one marker panel were studied in multiple independent populations. Hypermethylation of the markers RASSF1, BRCA, PITX2, CDH1, RARB, PCDH10 and PGR, and the marker panel GSTP1, RASSF1 and RARB showed a statistically significant correlation with poor disease outcome that was confirmed in at least one other, independent study.
Conclusion: This systematic review provides an overview on published prognostic DNA methylation markers for hormone receptor-positive breast cancer and identifies eight markers that have been independently validated. Analysis of the reporting quality of included studies suggests that future research on this topic would benefit from standardised reporting guidelines.
Keywords: Biomarkers; Breast cancer; DNA methylation; Hormone receptor positive; Luminal breast cancer; Oestrogen receptor positive; Prognosis; Promoter CpG island methylation; Survival.
Conflict of interest statement
Prof. dr. Tjan-Heijnen reports grants from AstraZeneca, during the conduct of the study; grants and non-financial support from Roche; grants and non-financial support from Pfizer; grants and non-financial support from Novartis; grants and non-financial support from E Lilly; and grants from Eisai, outside the submitted work. The other authors declare that they have no competing interests.
Figures

Similar articles
-
Prognostic role of methylated GSTP1, p16, ESR1 and PITX2 in patients with breast cancer: A systematic meta-analysis under the guideline of PRISMA.Medicine (Baltimore). 2017 Jul;96(28):e7476. doi: 10.1097/MD.0000000000007476. Medicine (Baltimore). 2017. PMID: 28700487 Free PMC article.
-
Prognostic relevance of promoter hypermethylation of multiple genes in breast cancer patients.Cell Oncol. 2009;31(6):487-500. doi: 10.3233/CLO-2009-0507. Cell Oncol. 2009. PMID: 19940364 Free PMC article.
-
Prognostic DNA methylation markers for sporadic colorectal cancer: a systematic review.Clin Epigenetics. 2018 Mar 14;10:35. doi: 10.1186/s13148-018-0461-8. eCollection 2018. Clin Epigenetics. 2018. PMID: 29564023 Free PMC article.
-
DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy.Clin Cancer Res. 2009 Jan 1;15(1):315-23. doi: 10.1158/1078-0432.CCR-08-0166. Clin Cancer Res. 2009. PMID: 19118060 Clinical Trial.
-
Clinicopathological significance and potential drug target of CDH1 in breast cancer: a meta-analysis and literature review.Drug Des Devel Ther. 2015 Sep 18;9:5277-85. doi: 10.2147/DDDT.S86929. eCollection 2015. Drug Des Devel Ther. 2015. PMID: 26425077 Free PMC article. Review.
Cited by
-
Identification of four novel hub genes as monitoring biomarkers for colorectal cancer.Hereditas. 2022 Jan 29;159(1):11. doi: 10.1186/s41065-021-00216-7. Hereditas. 2022. PMID: 35093172 Free PMC article.
-
Epigenetic Therapies and Biomarkers in Breast Cancer.Cancers (Basel). 2022 Jan 18;14(3):474. doi: 10.3390/cancers14030474. Cancers (Basel). 2022. PMID: 35158742 Free PMC article. Review.
-
Automated and rapid detection of cancer in suspicious axillary lymph nodes in patients with breast cancer.NPJ Breast Cancer. 2021 Jul 7;7(1):89. doi: 10.1038/s41523-021-00298-6. NPJ Breast Cancer. 2021. PMID: 34234148 Free PMC article.
-
Circulating Tumor DNA Is a Variant of Liquid Biopsy with Predictive and Prognostic Clinical Value in Breast Cancer Patients.Int J Mol Sci. 2023 Dec 2;24(23):17073. doi: 10.3390/ijms242317073. Int J Mol Sci. 2023. PMID: 38069396 Free PMC article. Review.
-
Selective Estrogen Receptor Modulators' (SERMs) Influence on TET3 Expression in Breast Cancer Cell Lines with Distinct Biological Subtypes.Int J Mol Sci. 2024 Aug 6;25(16):8561. doi: 10.3390/ijms25168561. Int J Mol Sci. 2024. PMID: 39201247 Free PMC article.
References
-
- American Cancer society . Global cancer facts & figures. 3. Atlanta: American Cancer Society; 2015.
-
- Peto R, Davies C, Godwin J, Gray R, Pan HC, Clarke M, et al. Comparisons between different polychemotherapy regimens for early breast cancer: meta-analyses of long-term outcome among 100,000 women in 123 randomised trials. Lancet. 2012;379(9814):432–444. doi: 10.1016/S0140-6736(11)61625-5. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

