The Role of pkc-3 and Genetic Suppressors in Caenorhabditis elegans Epithelial Cell Junction Formation
- PMID: 32005655
- PMCID: PMC7153940
- DOI: 10.1534/genetics.120.303085
The Role of pkc-3 and Genetic Suppressors in Caenorhabditis elegans Epithelial Cell Junction Formation
Abstract
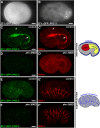
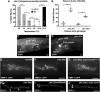
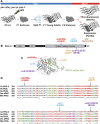
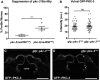
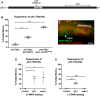
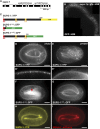
Epithelial cells form intercellular junctions to strengthen cell-cell adhesion and limit diffusion, allowing epithelia to function as dynamic tissues and barriers separating internal and external environments. Junctions form as epithelial cells differentiate; clusters of junction proteins first concentrate apically, then mature into continuous junctional belts that encircle and connect each cell. In mammals and Drosophila, atypical protein kinase C (aPKC) is required for junction maturation, although how it contributes to this process is poorly understood. A role for the Caenorhabditis elegans aPKC homolog PKC-3 in junction formation has not been described previously. Here, we show that PKC-3 is essential for junction maturation as epithelia first differentiate. Using a temperature-sensitive allele of pkc-3 that causes junction breaks in the spermatheca and leads to sterility, we identify intragenic and extragenic suppressors that render pkc-3 mutants fertile. Intragenic suppressors include an unanticipated stop-to-stop mutation in the pkc-3 gene, providing evidence for the importance of stop codon identity in gene activity. One extragenic pkc-3 suppressor is a loss-of-function allele of the lethal(2) giant larvae homolog lgl-1, which antagonizes aPKC within epithelia of Drosophila and mammals, but was not known previously to function in C. elegans epithelia. Finally, two extragenic suppressors are loss-of-function alleles of sups-1-a previously uncharacterized gene. We show that SUPS-1 is an apical extracellular matrix protein expressed in epidermal cells, suggesting that it nonautonomously regulates junction formation in the spermatheca. These findings establish a foundation for dissecting the role of PKC-3 and interacting genes in epithelial junction maturation.
Keywords: aPKC; adherens junction; cell polarity; kinase; stop codon; suppressor.
Copyright © 2020 by the Genetics Society of America.
Figures
Similar articles
-
PAR-6 is required for junction formation but not apicobasal polarization in C. elegans embryonic epithelial cells.Development. 2007 Apr;134(7):1259-68. doi: 10.1242/dev.02833. Epub 2007 Feb 21. Development. 2007. PMID: 17314130
-
Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures.J Cell Biol. 2001 Mar 19;152(6):1183-96. doi: 10.1083/jcb.152.6.1183. J Cell Biol. 2001. PMID: 11257119 Free PMC article.
-
PAR-3 mediates the initial clustering and apical localization of junction and polarity proteins during C. elegans intestinal epithelial cell polarization.Development. 2010 Jun;137(11):1833-42. doi: 10.1242/dev.047647. Epub 2010 Apr 28. Development. 2010. PMID: 20431121 Free PMC article.
-
Role of Lgl/Dlg/Scribble in the regulation of epithelial junction, polarity and growth.Front Biosci. 2008 May 1;13:6693-707. doi: 10.2741/3182. Front Biosci. 2008. PMID: 18508688 Review.
-
Protein kinase C isotypes in C. elegans.J Biochem. 2002 Oct;132(4):519-22. doi: 10.1093/oxfordjournals.jbchem.a003251. J Biochem. 2002. PMID: 12359064 Review.
Cited by
-
Apical PAR complex proteins protect against programmed epithelial assaults to create a continuous and functional intestinal lumen.Elife. 2021 Jun 17;10:e64437. doi: 10.7554/eLife.64437. Elife. 2021. PMID: 34137371 Free PMC article.
-
A polarity pathway for exocyst-dependent intracellular tube extension.Elife. 2021 Mar 9;10:e65169. doi: 10.7554/eLife.65169. Elife. 2021. PMID: 33687331 Free PMC article.
-
Separable mechanisms drive local and global polarity establishment in the Caenorhabditiselegans intestinal epithelium.Development. 2022 Nov 15;149(22):dev200325. doi: 10.1242/dev.200325. Epub 2022 Nov 16. Development. 2022. PMID: 36264257 Free PMC article.
-
Whole genome sequencing facilitates intragenic variant interpretation following modifier screening in C. elegans.BMC Genomics. 2021 Nov 13;22(1):820. doi: 10.1186/s12864-021-08142-8. BMC Genomics. 2021. PMID: 34773966 Free PMC article.
-
C. elegans Afadin is required for epidermal morphogenesis and functionally interfaces with the cadherin-catenin complex and RhoGAP PAC-1/ARHGAP21.Dev Biol. 2024 Jul;511:12-25. doi: 10.1016/j.ydbio.2024.03.007. Epub 2024 Mar 29. Dev Biol. 2024. PMID: 38556137 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

