Functional analysis of Clostridium difficile sortase B reveals key residues for catalytic activity and substrate specificity
- PMID: 32005667
- PMCID: PMC7076211
- DOI: 10.1074/jbc.RA119.011322
Functional analysis of Clostridium difficile sortase B reveals key residues for catalytic activity and substrate specificity
Abstract
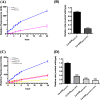
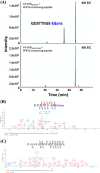
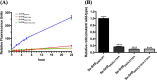
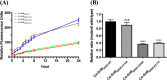
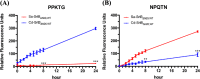
Most of Gram-positive bacteria anchor surface proteins to the peptidoglycan cell wall by sortase, a cysteine transpeptidase that targets proteins displaying a cell wall sorting signal. Unlike other bacteria, Clostridium difficile, the major human pathogen responsible for antibiotic-associated diarrhea, has only a single functional sortase (SrtB). Sortase's vital importance in bacterial virulence has been long recognized, and C. difficile sortase B (Cd-SrtB) has become an attractive therapeutic target for managing C. difficile infection. A better understanding of the molecular activity of Cd-SrtB may help spur the development of effective agents against C. difficile infection. In this study, using site-directed mutagenesis, biochemical and biophysical tools, LC-MS/MS, and crystallographic analyses, we identified key residues essential for Cd-SrtB catalysis and substrate recognition. To the best of our knowledge, we report the first evidence that a conserved serine residue near the active site participates in the catalytic activity of Cd-SrtB and also SrtB from Staphylococcus aureus The serine residue indispensable for SrtB activity may be involved in stabilizing a thioacyl-enzyme intermediate because it is neither a nucleophilic residue nor a substrate-interacting residue, based on the LC-MS/MS data and available structural models of SrtB-substrate complexes. Furthermore, we also demonstrated that residues 163-168 located on the β6/β7 loop of Cd-SrtB dominate specific recognition of the peptide substrate PPKTG. The results of this work reveal key residues with roles in catalysis and substrate specificity of Cd-SrtB.
Keywords: Clostridium difficile; crystal structure; cysteine transpeptidase; enzyme catalysis; fluorescence resonance energy transfer (FRET); protein chemistry; protein purification; protein sorting; protein structure; sortase B; substrate specificity.
© 2020 Kang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures



References
-
- Peery A. F., Dellon E. S., Lund J., Crockett S. D., McGowan C. E., Bulsiewicz W. J., Gangarosa L. M., Thiny M. T., Stizenberg K., Morgan D. R., Ringel Y., Kim H. P., DiBonaventura M. D., Carroll C. F., Allen J. K., et al. (2012) Burden of gastrointestinal disease in the United States: 2012 update. Gastroenterology 143, 1179–1187.e3 10.1053/j.gastro.2012.08.002 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

