Deciphering the Bifidobacterial Populations within the Canine and Feline Gut Microbiota
- PMID: 32005736
- PMCID: PMC7082561
- DOI: 10.1128/AEM.02875-19
Deciphering the Bifidobacterial Populations within the Canine and Feline Gut Microbiota
Abstract
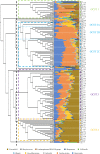
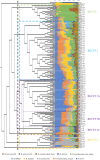
During the course of evolution, dogs and cats have been subjected to extensive domestication, becoming the principal companion animals for humans. For this reason, their health care, including their intestinal microbiota, is considered of considerable importance. However, the canine and feline gut microbiota still represent a largely unexplored research area. In the present work, we profiled the microbiota of 23 feline fecal samples by 16S rRNA gene and bifidobacterial internally transcribed spacer (ITS) approaches and compared this information with previously reported data from 138 canine fecal samples. The obtained data allowed the reconstruction of the core gut microbiota of the above-mentioned samples coupled with their classification into distinct community state types at both genus and species levels, identifying Bacteroides, Fusobacterium, and Prevotella 9 as the main bacterial components of the canine and feline gut microbiota. At the species level, the intestinal bifidobacterial gut communities of dogs and cats differed in terms of both species number and composition, as emphasized by a covariance analysis. Together, our findings show that the intestinal populations of cats and dogs are similar in terms of genus-level taxonomical composition, while at the bifidobacterial species level, clear differences were observed, indicative of host-specific colonization behavior by particular bifidobacterial taxa.IMPORTANCE Currently, domesticated dogs and cats are the most cherished companion animals for humans, and concerns about their health and well-being are therefore important. In this context, the gut microbiota plays a crucial role in maintaining and promoting host health. However, despite the social relevance of domesticated dogs and cats, their intestinal microbial communities are still far from being completely understood. In this study, the taxonomical composition of canine and feline gut microbiota was explored at genus and bifidobacterial species levels, allowing classification of these microbial populations into distinct gut community state types at either of the two investigated taxonomic levels. Furthermore, the reconstruction of core gut microbiota coupled with covariance network analysis based on bifidobacterial internally transcribed spacer (ITS) profiling revealed differences in the bifidobacterial compositions of canine and feline gut microbiota, suggesting that particular bifidobacterial species have developed a selective ability to colonize a specific host.
Keywords: bifidobacteria; cat; dog; gut microbiota; metagenomics.
Copyright © 2020 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

