A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration
- PMID: 32005911
- PMCID: PMC6994629
- DOI: 10.1038/s41598-020-58510-9
A transcriptome-wide association study based on 27 tissues identifies 106 genes potentially relevant for disease pathology in age-related macular degeneration
Abstract
Genome-wide association studies (GWAS) for late stage age-related macular degeneration (AMD) have identified 52 independent genetic variants with genome-wide significance at 34 genomic loci. Typically, such an approach rarely results in the identification of functional variants implicating a defined gene in the disease process. We now performed a transcriptome-wide association study (TWAS) allowing the prediction of effects of AMD-associated genetic variants on gene expression. The TWAS was based on the genotypes of 16,144 late-stage AMD cases and 17,832 healthy controls, and gene expression was imputed for 27 different human tissues which were obtained from 134 to 421 individuals. A linear regression model including each individuals imputed gene expression data and the respective AMD status identified 106 genes significantly associated to AMD variants in at least one tissue (Q-value < 0.001). Gene enrichment analysis highlighted rather systemic than tissue- or cell-specific processes. Remarkably, 31 of the 106 genes overlapped with significant GWAS signals of other complex traits and diseases, such as neurological or autoimmune conditions. Taken together, our study highlights the fact that expression of genes associated with AMD is not restricted to retinal tissue as could be expected for an eye disease of the posterior pole, but instead is rather ubiquitous suggesting processes underlying AMD pathology to be of systemic nature.
Conflict of interest statement
The authors declare no competing interests.
Figures


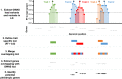
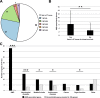
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

