Regulatory R-loops as facilitators of gene expression and genome stability
- PMID: 32005969
- PMCID: PMC7116639
- DOI: 10.1038/s41580-019-0206-3
Regulatory R-loops as facilitators of gene expression and genome stability
Abstract
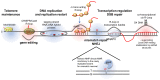
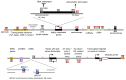
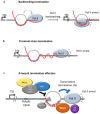
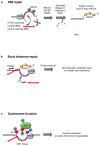
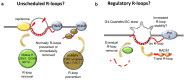
R-loops are three-stranded structures that harbour an RNA-DNA hybrid and frequently form during transcription. R-loop misregulation is associated with DNA damage, transcription elongation defects, hyper-recombination and genome instability. In contrast to such 'unscheduled' R-loops, evidence is mounting that cells harness the presence of RNA-DNA hybrids in scheduled, 'regulatory' R-loops to promote DNA transactions, including transcription termination and other steps of gene regulation, telomere stability and DNA repair. R-loops formed by cellular RNAs can regulate histone post-translational modification and may be recognized by dedicated reader proteins. The two-faced nature of R-loops implies that their formation, location and timely removal must be tightly regulated. In this Perspective, we discuss the cellular processes that regulatory R-loops modulate, the regulation of R-loops and the potential differences that may exist between regulatory R-loops and unscheduled R-loops.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

