MINI SEED 2 (MIS2) Encodes a Receptor-like Kinase that Controls Grain Size and Shape in Rice
- PMID: 32006119
- PMCID: PMC6994593
- DOI: 10.1186/s12284-020-0368-9
MINI SEED 2 (MIS2) Encodes a Receptor-like Kinase that Controls Grain Size and Shape in Rice
Abstract
Background: Grain size is a key agronomic trait that is directly associated with grain yield in rice. Although several genes related to grain size in rice have been identified, our understanding of the mechanism of grain development is still limited.
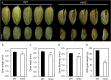
Results: In this study, we reported the characterization of a novel seed size mutant mini seed 2 (mis2), in which the grain showed reduced length, width and thickness along with wrinkled surface. Microscopic analysis revealed that the spikelet epidermal cell size was reduced but the cell number was increased in the mis2 mutant, suggesting that MIS2 controls grain size by coordinately regulating epidermal cell size and cell number. Map-based cloning revealed that MIS2 encodes a receptor-like kinase CRINKLY4 (CR4) which showed the highest expression in developing panicles. The MIS2 protein is localized primarily on the plasma membrane along with the endosome. However, the Arg258Gln mutation located in extracellular domain in the mis2 mutant disturbed its subcellular localization. Additionally, three major haplotypes of MIS2 were identified in the japonica, indica and aus rice cultivars. The 18-bp InDel (insertion and deletion) in the 5'-UTR (untranslated region) caused different expression level of MIS2 in haplotypes.
Conclusions: We reported a key role of OsCR4 in controlling grain size and shape by coordinately regulating epidermal cell size and cell number. The Arg258 in the extracellular seven-repeat domain is essential for the correct subcellular behavior and function of the OsCR4 protein.
Keywords: CRINKLY4; Epidermal cells; Grain size and shape; MIS2; OsCR4; Receptor-like kinase; Rice; Spikelets.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Silencing of an Ubiquitin Ligase Increases Grain Width and Weight in indica Rice.Front Genet. 2021 Jan 12;11:600378. doi: 10.3389/fgene.2020.600378. eCollection 2020. Front Genet. 2021. PMID: 33510769 Free PMC article.
-
Natural Variation in the Promoter of GSE5 Contributes to Grain Size Diversity in Rice.Mol Plant. 2017 May 1;10(5):685-694. doi: 10.1016/j.molp.2017.03.009. Epub 2017 Mar 30. Mol Plant. 2017. PMID: 28366824
-
OsHXK3 encodes a hexokinase-like protein that positively regulates grain size in rice.Theor Appl Genet. 2022 Oct;135(10):3417-3431. doi: 10.1007/s00122-022-04189-7. Epub 2022 Aug 8. Theor Appl Genet. 2022. PMID: 35941236
-
Control of grain size in rice.Plant Reprod. 2018 Sep;31(3):237-251. doi: 10.1007/s00497-018-0333-6. Epub 2018 Mar 10. Plant Reprod. 2018. PMID: 29523952 Review.
-
Genes and Their Molecular Functions Determining Seed Structure, Components, and Quality of Rice.Rice (N Y). 2022 Mar 18;15(1):18. doi: 10.1186/s12284-022-00562-8. Rice (N Y). 2022. PMID: 35303197 Free PMC article. Review.
Cited by
-
Unveiling the genetic architecture for lodging resistance in rice (Oryza sativa. L) by genome-wide association analyses.Front Genet. 2022 Sep 6;13:960007. doi: 10.3389/fgene.2022.960007. eCollection 2022. Front Genet. 2022. PMID: 36147492 Free PMC article.
-
Comprehensive Transcriptome Analyses Reveal Candidate Genes for Variation in Seed Size/Weight During Peanut (Arachis hypogaea L.) Domestication.Front Plant Sci. 2021 May 19;12:666483. doi: 10.3389/fpls.2021.666483. eCollection 2021. Front Plant Sci. 2021. PMID: 34093624 Free PMC article.
-
Positional cloning identified HvTUBULIN8 as the candidate gene for round lateral spikelet (RLS) in barley (Hordeum vulgare L.).Theor Appl Genet. 2023 Jan;136(1):7. doi: 10.1007/s00122-023-04272-7. Epub 2023 Jan 19. Theor Appl Genet. 2023. PMID: 36656367 Free PMC article.
-
Comparative transcriptome analysis, unfolding the pathways regulating the seed-size trait in cultivated lentil (Lens culinaris Medik.).Front Genet. 2022 Aug 10;13:942079. doi: 10.3389/fgene.2022.942079. eCollection 2022. Front Genet. 2022. PMID: 36035144 Free PMC article.
-
Comprehensive Comparative Analysis of the GATA Transcription Factors in Four Rosaceae Species and Phytohormonal Response in Chinese Pear (Pyrus bretschneideri) Fruit.Int J Mol Sci. 2021 Nov 19;22(22):12492. doi: 10.3390/ijms222212492. Int J Mol Sci. 2021. PMID: 34830372 Free PMC article.
References
-
- Ahmadi N, Audebert A, Bennett MJ, Bishopp A, de Oliveira AC, Courtois B, Diedhiou A, Diévart A, Gantet P, Ghesquière A, Guiderdoni E, Henry A, Inukai Y, Kochian L, Laplaze L, Lucas M, Luu DT, Manneh B, Mo X, Muthurajan R, Périn C, Price A, Robin S, Sentenac H, Sine B, Uga Y, Véry AA, Wissuwa M, Wu P, Xu J. The roots of future rice harvests. Rice. 2014;7:29. doi: 10.1186/s12284-014-0029-y. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

