Multiplex bacterial polymerase chain reaction in a cohort of patients with pleural effusion
- PMID: 32007106
- PMCID: PMC6995182
- DOI: 10.1186/s12879-020-4793-6
Multiplex bacterial polymerase chain reaction in a cohort of patients with pleural effusion
Abstract
Background: The identification of the pathogens in pleural effusion has mainly relied on conventional bacterial culture or single species polymerase chain reaction (PCR), both with relatively low sensitivity. We investigated the efficacy of a commercially available multiplex bacterial PCR assay developed for pneumonia to identify the pathogens involved in pleural infection, particularly empyema.
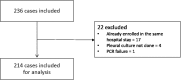
Methods: A prospective, monocentric, observational study including 194 patients with pleural effusion. Patients were evaluated based on imaging, laboratory values, pleura ultrasound and results of thoracentesis including conventional microbiology studies during hospitalisation. Multiplex bacterial PCR (Curetis Unyvero p55) was performed in batch and had no influence on therapeutic decisions.
Results: Overall, there were 51/197 cases with transudate and 146/197 with exudate. In 42% (n = 90/214) there was a clinical suspicion of parapneumonic effusion and the final clinical diagnosis of empyema was made in 29% (n = 61/214) of all cases. The most common microorganisms identified in the cases diagnosed with empyema were anaerobes [31] followed by gram-positive cocci [10] and gram-negative rods [4]. The multiplex PCR assay identified more of the pathogens on the panel than the conventional methods (23.3% (7/30) vs. 6.7% (2/30), p = 0.008).
Conclusion: The multiplex PCR-based assay had a higher sensitivity and specificity than conventional microbiology when only the pathogens on the pneumonia panel were taken into account. A dedicated pleural empyema multiplex PCR panel including anaerobes would be needed to cover most common pathogens involved in pleural infection.
Keywords: Bacteria; Multiplex PCR; Para-pneumonic effusion; Pathogen; Pleural empyema.
Conflict of interest statement
The authors declare that they have no competing interests. The Curetis Unyvero P55 assay cartridges were donated by Curetis AG, Holzgerlingen, Germany. The sponsors of this investigator-initiated project had no involvement in the design and conduct of the study, collection, management, analysis, and interpretation of the data, nor in the preparation, review, and approval of the manuscript or decision to submit the manuscript.
Figures
Similar articles
-
Survey of childhood empyema in Asia: implications for detecting the unmeasured burden of culture-negative bacterial disease.BMC Infect Dis. 2008 Jul 11;8:90. doi: 10.1186/1471-2334-8-90. BMC Infect Dis. 2008. PMID: 18620553 Free PMC article.
-
Etiology of parapneumonic effusion and pleural empyema in children. The role of conventional and molecular microbiological tests.Respir Med. 2016 Jul;116:28-33. doi: 10.1016/j.rmed.2016.05.009. Epub 2016 May 10. Respir Med. 2016. PMID: 27296817 Free PMC article.
-
A Retrospective Analysis of Anaerobic Bacteria Isolated in 236 Cases of Pleural Empyema and their Prevalance of Antimicrobial Resistance in Turkey.Clin Lab. 2018 Jul 1;64(7):1269-1277. doi: 10.7754/Clin.Lab.2018.180317. Clin Lab. 2018. PMID: 30146848
-
Antibiotic treatment of patients with pneumonia and pleural effusion.Curr Opin Pulm Med. 1998 Jul;4(4):230-4. doi: 10.1097/00063198-199807000-00009. Curr Opin Pulm Med. 1998. PMID: 10813239 Review.
-
[From pneumonic infiltration to parapneumonic effusion--from effusion to pleural empyema: internal medicine aspects of parapneumonic effusion development and pleural empyema].Wien Med Wochenschr. 2003;153(15-16):349-53. doi: 10.1007/s10354-003-0008-1. Wien Med Wochenschr. 2003. PMID: 13677257 Review. German.
Cited by
-
GenomicGapID: leveraging spatial distribution of conserved genomic sites for broad-spectrum microbial identification.Microbiol Spectr. 2025 May 6;13(5):e0281724. doi: 10.1128/spectrum.02817-24. Epub 2025 Mar 14. Microbiol Spectr. 2025. PMID: 40084960 Free PMC article.
-
Identification of Microbiome Etiology Associated With Drug Resistance in Pleural Empyema.Front Cell Infect Microbiol. 2021 Mar 16;11:637018. doi: 10.3389/fcimb.2021.637018. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33796482 Free PMC article.
-
Development of a Rapid, Culture-Free, Universal Microbial Identification System Using Internal Transcribed Spacer Targeting Primers.J Infect Dis. 2025 Jun 2;231(5):1155-1164. doi: 10.1093/infdis/jiae545. J Infect Dis. 2025. PMID: 39503259 Free PMC article.
-
Assessment of Intraoperative Microbiological Culture in Patients with Empyema: Comparison with Preoperative Microbiological Culture.Ann Thorac Cardiovasc Surg. 2021 Dec 20;27(6):346-354. doi: 10.5761/atcs.oa.20-00327. Epub 2021 May 8. Ann Thorac Cardiovasc Surg. 2021. PMID: 33967122 Free PMC article.
-
Use of multiplex PCR in pleural effusion: is it necessary to change the paradigm of culture-based methods?Access Microbiol. 2023 Nov 15;5(11):000612.v3. doi: 10.1099/acmi.0.000612.v3. eCollection 2023. Access Microbiol. 2023. PMID: 38074107 Free PMC article.
References
-
- Le Monnier A, Carbonnelle E, Zahar J-R, Le Bourgeois M, Abachin E, Quesne G, et al. Microbiological diagnosis of empyema in children: comparative evaluations by culture, polymerase chain reaction, and pneumococcal antigen detection in pleural fluids. Clin Infect Dis. 2006;42(8):1135–1140. doi: 10.1086/502680. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources