Deep learning-based clustering approaches for bioinformatics
- PMID: 32008043
- PMCID: PMC7820885
- DOI: 10.1093/bib/bbz170
Deep learning-based clustering approaches for bioinformatics
Abstract
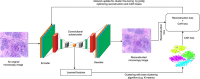
Clustering is central to many data-driven bioinformatics research and serves a powerful computational method. In particular, clustering helps at analyzing unstructured and high-dimensional data in the form of sequences, expressions, texts and images. Further, clustering is used to gain insights into biological processes in the genomics level, e.g. clustering of gene expressions provides insights on the natural structure inherent in the data, understanding gene functions, cellular processes, subtypes of cells and understanding gene regulations. Subsequently, clustering approaches, including hierarchical, centroid-based, distribution-based, density-based and self-organizing maps, have long been studied and used in classical machine learning settings. In contrast, deep learning (DL)-based representation and feature learning for clustering have not been reviewed and employed extensively. Since the quality of clustering is not only dependent on the distribution of data points but also on the learned representation, deep neural networks can be effective means to transform mappings from a high-dimensional data space into a lower-dimensional feature space, leading to improved clustering results. In this paper, we review state-of-the-art DL-based approaches for cluster analysis that are based on representation learning, which we hope to be useful, particularly for bioinformatics research. Further, we explore in detail the training procedures of DL-based clustering algorithms, point out different clustering quality metrics and evaluate several DL-based approaches on three bioinformatics use cases, including bioimaging, cancer genomics and biomedical text mining. We believe this review and the evaluation results will provide valuable insights and serve a starting point for researchers wanting to apply DL-based unsupervised methods to solve emerging bioinformatics research problems.
© The Author(s) 2020. Published by Oxford University Press.
Figures





References
-
- Min E, Guo X, Qiang, et al. A survey of clustering with deep learning: from the perspective of network architecture. IEEE Access 2018; 6:39501–14.
-
- Gan G, Ma C, Wu J. Data clustering: theory, algorithms, and applications, Vol. 20 SIAM, 2007.
-
- Costa IG, de Carvalho F d AT, de Souto MCP. Comparative analysis of clustering methods for gene expression time course data. Genet Mol Biol 2004; 27(4): 623–31.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

