The Deep Genome Project
- PMID: 32008577
- PMCID: PMC6996159
- DOI: 10.1186/s13059-020-1931-9
The Deep Genome Project
Conflict of interest statement
PF is a member of the Scientific Advisory Boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd. The other authors declare that they have no competing interests.
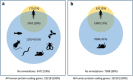
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- G9521010/MRC_/Medical Research Council/United Kingdom
- MC_PC_14089/MRC_/Medical Research Council/United Kingdom
- MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- UM1 OD023221/OD/NIH HHS/United States
- UM1 HG006348/HG/NHGRI NIH HHS/United States
- MC_EX_MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- MC_U142684172/MRC_/Medical Research Council/United Kingdom
- UM1 HG006370/HG/NHGRI NIH HHS/United States
- MC_U142684171/MRC_/Medical Research Council/United Kingdom
- FS/12/82/29736/BHF_/British Heart Foundation/United Kingdom
- MC_UP_1502/1/MRC_/Medical Research Council/United Kingdom
- UM1 OD023222/OD/NIH HHS/United States

