State-Wide Genomic and Epidemiological Analyses of Vancomycin-Resistant Enterococcus faecium in Tasmania's Public Hospitals
- PMID: 32010070
- PMCID: PMC6975128
- DOI: 10.3389/fmicb.2019.02940
State-Wide Genomic and Epidemiological Analyses of Vancomycin-Resistant Enterococcus faecium in Tasmania's Public Hospitals
Abstract
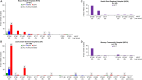
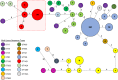
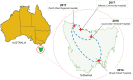
From 2015 onwards, the number of vancomycin-resistant Enterococcus faecium (VREfm) isolates increased in Tasmania. Previously, we examined the transmission of VREfm at the Royal Hobart Hospital (RHH). In this study, we performed a state-wide analysis of VREfm from Tasmania's four public acute hospitals. Whole-genome analysis was performed on 331 isolates collected from screening and clinical specimens of VREfm. In silico multi-locus sequence typing (MLST) was used to determine the relative abundance of broad sequence types (ST) across the state. Core genome MLST (cgMLST) was then applied to identify potential clades within the ST groupings followed by single-nucleotide polymorphic (SNP) analysis. This work revealed that differences in VREfm profiles are evident between the state's two largest hospitals with the dominant vanA types being ST80 at the RHH and ST1421 at Launceston General Hospital (LGH). A higher number of VREfm cases were recorded at LGH (n = 54 clinical, n = 122 colonization) compared to the RHH (n = 14 clinical, n = 67 colonization) during the same time period, 2014-2016. Eleven of the clinical isolates from LGH were vanA and belonged to ST1421 (n = 8), ST1489 (n = 1), ST233 (n = 1), and ST80 (n = 1) whereas none of the clinical isolates from the RHH were vanA. For the recently described ST1421, cgMLST established the presence of individual clusters within this sequence type that were common to more than one hospital and that included isolates with a low amount of SNP variance (≤16 SNPs). A spatio-temporal analysis revealed that VREfm vanA ST1421 was first detected at the RHH in 2014 and an isolate belonging to the same cgMLST cluster was later collected at LGH in 2016. Inclusion of isolates from two smaller hospitals, the North West Regional Hospital (NRH) and the Mersey Community Hospital (MCH) found that ST1421 was present in both of these institutions in 2017. These findings illustrate the spread of a recently described sequence type of VREfm, ST1421, to multiple hospitals in an Australian state within a relatively short time span.
Keywords: Enterococcus faecium; multi-locus sequence typing; single nucleotide polymorphism; vancomycin; whole genome sequencing.
Copyright © 2020 Leong, Kalukottege, Cooley, Anderson, Wells, Langford and O’Toole.
Figures





References
-
- Act Parliamentary Counsel (2016). Public Health Act 1997, ed. Health M. F. (Canberra: ACT; )
-
- Andersson P., Beckingham W., Gorrie C. L., Kennedy K., Daveson K., Ballard S. A., et al. (2019). Vancomycin-resistant Enterococcus (VRE) outbreak in a neonatal intensive care unit and special care nursery at a tertiary-care hospital in Australia-A retrospective case-control study. Infect. Control Hosp. Epidemiol. 40 551–558. 10.1017/ice.2019.41 - DOI - PubMed
-
- Andrews S., Krueger F., Segonds-Pichon A., Biggins L., Krueger C., Wingett S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham: the Brabaham Institute.
-
- Australian Commission on Safety and Quality in Health Care (ACSQHC) (2017). AURA 2017: Second Australian Report on Antimicrobial Use, and Resistance. in Human Health. Sydney: ACSQHC.
LinkOut - more resources
Full Text Sources

