Investigation of Genetic Relationships Between Hanseniaspora Species Found in Grape Musts Revealed Interspecific Hybrids With Dynamic Genome Structures
- PMID: 32010076
- PMCID: PMC6974558
- DOI: 10.3389/fmicb.2019.02960
Investigation of Genetic Relationships Between Hanseniaspora Species Found in Grape Musts Revealed Interspecific Hybrids With Dynamic Genome Structures
Abstract
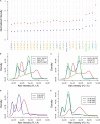
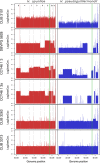
Hanseniaspora, a predominant yeast genus of grape musts, includes sister species recently reported as fast evolving. The aim of this study was to investigate the genetic relationships between the four most closely related species, at the population level. A multi-locus sequence typing strategy based on five markers was applied on 107 strains, confirming the clear delineation of species H. uvarum, H. opuntiae, H. guilliermondii, and H. pseudoguilliermondii. Huge variations were observed in the level of intraspecific nucleotide diversity, and differences in heterozygosity between species indicate different life styles. No clear population structure was detected based on geographical or substrate origins. Instead, H. guilliermondii strains clustered into two distinct groups, which may reflect a recent step toward speciation. Interspecific hybrids were detected between H. opuntiae and H. pseudoguilliermondii. Their characterization using flow cytometry, karyotypes and genome sequencing showed different genome structures in different ploidy contexts: allodiploids, allotriploids, and allotetraploids. Subculturing of an allotriploid strain revealed chromosome loss equivalent to one chromosome set, followed by an auto-diploidization event, whereas another auto-diploidized tetraploid showed a segmental duplication. Altogether, these results suggest that Hanseniaspora genomes are not only fast evolving but also highly dynamic.
Keywords: Hanseniaspora guilliermondii; Hanseniaspora uvarum; MLST; biodiversity; evolution; yeast.
Copyright © 2020 Saubin, Devillers, Proust, Brier, Grondin, Pradal, Legras and Neuvéglise.
Figures




References
-
- Bizzarri M., Cassanelli S., Bartolini L., Pryszcz L. P., Duskova M., Sychrova H., et al. (2019). Interplay of chimeric mating-type loci impairs fertility rescue and accounts for intra-strain variability in zygosaccharomyces rouxii interspecies Hybrid ATCC42981. Front. Genet. 10:137. 10.3389/fgene.2019.00137 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

