Analysis of an Intrinsic Mycovirus Associated With Reduced Virulence of the Human Pathogenic Fungus Aspergillus fumigatus
- PMID: 32010101
- PMCID: PMC6978690
- DOI: 10.3389/fmicb.2019.03045
Analysis of an Intrinsic Mycovirus Associated With Reduced Virulence of the Human Pathogenic Fungus Aspergillus fumigatus
Abstract
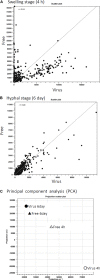
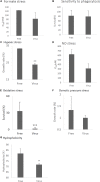
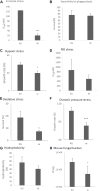
Aspergillus fumigatus is an airborne fungal pathogen that causes severe infections with invasive growth in immunocompromised patients. Several mycoviruses have recently been isolated from A. fumigatus strains, but there are presently no reports of mycoviral-mediated reduction or elimination of fungal pathogenicity in vertebrate models. Here, we report the biological features of a novel mycovirus, A. fumigatus chrysovirus 41362 (AfuCV41362), isolated from the hypovirulent A. fumigatus strain IFM 41362. The AfuCV41362 genome is comprised of four dsRNAs, each of which contains a single ORF (ORF1-4). ORF1 encodes a protein with sequence similarity to RNA-dependent RNA polymerases of viruses in the family Chrysoviridae, while ORF3 encodes a putative capsid protein. Viral RNAs are expressed primarily during the germination stage, and RNA-seq analysis of virus-infected A. fumigatus at the germination stage suggested that the virus suppressed expression of several pathogenicity-associated host genes, including hypoxia adaptation and nitric oxide detoxification genes. In vitro functional analysis revealed that the virus-infected strain had reduced tolerance to environmental stressors. Virus-infected A. fumigatus strain IFM 41362 had reduced virulence in vivo compared to the virus-free strain in a mouse infection model. Furthermore, introduction of the mycovirus to a natively virus-free KU A. fumigatus strain induced virus-infected phenotypes. To identify mycovirus genes responsible for the reduced virulence of A. fumigatus, each viral ORF was ectopically expressed in the virus-free KU strain. Ectopic expression of the individual ORFs only nominally reduced virulence of the host fungus in a mouse infection model. However, we found that ORF3 and ORF4 reduced tolerance to environmental stresses in in vitro analysis. Based on these results, we suggest that the AfuCV41362 mycovirus ORF3 and ORF4 reduce fungal virulence by suppressing stress tolerance together with other viral genes, rather than alone.
Keywords: Aspergillus fumigatus; Chrysoviridae; dsRNA; hypovirulence; mycovirus.
Copyright © 2020 Takahashi-Nakaguchi, Shishido, Yahara, Urayama, Sakai, Chibana, Kamei, Moriyama and Gonoi.
Figures










References
LinkOut - more resources
Full Text Sources
Research Materials

