Molecular Evolution of the Fusion Protein (F) Gene in Human Respirovirus 3
- PMID: 32010105
- PMCID: PMC6974460
- DOI: 10.3389/fmicb.2019.03054
Molecular Evolution of the Fusion Protein (F) Gene in Human Respirovirus 3
Abstract
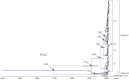
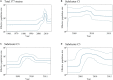
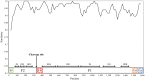
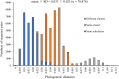
To elucidate the evolution of human respirovirus 3 (HRV3), we performed detailed genetic analyses of the F gene (full-length) detected from hundreds of HRV3 strains obtained from various geographic regions. First, we performed time-scaled evolutionary analyses using the Bayesian Markov chain Monte Carlo method. Then, we performed analyses of phylodynamics, similarity, phylogenetic distance, selective pressure, and conformational B-cell epitope with the F-protein structural analyses. Time-scaled phylogenetic tree showed that the common ancestor of HRV3 and bovine respirovirus 3 diverged over 300 years ago and subdivided it into three major clusters and four subclusters during the most recent 100 years. The overall evolutionary rate was approximately 10-3 substitutions/site/year. Indigenous similarity was seen in the present strains, and the mean phylogenetic distance were 0.033. Many negative selection sites were seen in the ectodomain. The conformational epitopes did not correspond to the neutralizing antibody binding sites. These results suggest that the HRV3 F gene is relatively conserved and restricted in this diversity to preserve the protein function, although these strains form many branches on the phylogenetic tree. Furthermore, HRV3 reinfection may be responsible for discordances between the conformational epitopes and the neutralizing antibody binding sites of the F protein. These findings contribute to a better understanding of HRV3 virology.
Keywords: conformational epitope; fusion peptide; fusion protein (F) gene; human respirovirus 3; molecular evolution.
Copyright © 2020 Aso, Kimura, Ishii, Saraya, Kurai, Matsushima, Nagasawa, Ryo and Takizawa.
Figures

References
-
- Abedi G. R., Prill M. M., Langley G. E., Wikswo M. E., Curns A. T., Schneider E. (2016). Estimates of parainfluenza virus-associated hospitalizations and cost among children aged less than 5 years in the United States, 1998-2010. J. Pediatric Infect. Dis. Soc. 5 7–13. 10.1093/jpids/piu047 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

