Proteomic Profiling Reveals the Architecture of Granulomatous Lesions Caused by Tuberculosis and Mycobacterium avium Complex Lung Disease
- PMID: 32010116
- PMCID: PMC6978656
- DOI: 10.3389/fmicb.2019.03081
Proteomic Profiling Reveals the Architecture of Granulomatous Lesions Caused by Tuberculosis and Mycobacterium avium Complex Lung Disease
Abstract
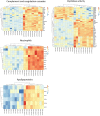
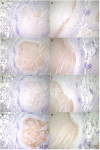
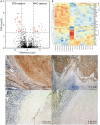
Tuberculosis (TB) and Mycobacterium avium complex lung disease (MAC-LD) are both characterized pathologically by granuloma lesions, which are typically composed of a necrotic caseum at the center surrounded by fibrotic cells and lymphocytes. Although the histological characterization of TB and MAC-LD granulomas has been well-documented, their molecular signatures have not been fully evaluated. In this research we applied mass spectrometry-based proteomics combined with laser microdissection to investigate the unique protein markers in human mycobacterial granulomatous lesions. Comparing the protein abundance between caseous and cellular sub-compartments of mycobacterial granulomas, we found distinct differences. Proteins involved in cellular metabolism in transcription and translation were abundant in cellular regions, while in caseous regions proteins related to antimicrobial response accumulated. To investigate the determinants of their heterogeneity, we compared the protein abundance in caseous regions between TB and MAC-LD granulomas. We found that several proteins were significantly abundant in the MAC-LD caseum of which proteomic profiles were different from those of the TB caseum. Immunohistochemistry demonstrated that one of these proteins, Angiogenin, specifically localized to the caseous regions of selected MAC-LD granulomas. We also detected peptides derived from mycobacterial proteins in the granulomas of both diseases. This study provides new insights into the architecture of granulomatous lesions in TB and MAC-LD.
Keywords: Mycobacterium avium complex lung disease; granuloma; necrotic caseum; proteomics; tuberculosis.
Copyright © 2020 Seto, Morimoto, Yoshida, Hiramatsu, Hijikata, Nagata, Kikuchi, Shiraishi, Kurashima and Keicho.
Figures





References
-
- Chitale S., Ehrt S., Kawamura I., Fujimura T., Shimono N., Anand, et al. (2001). Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell. Microbiol. 3 247–254. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

