Sequence conservation of mitochondrial (mt)DNA during expansion of clonal mammary epithelial populations suggests a common mtDNA template in CzechII mice
- PMID: 32010429
- PMCID: PMC6968779
- DOI: 10.18632/oncotarget.27429
Sequence conservation of mitochondrial (mt)DNA during expansion of clonal mammary epithelial populations suggests a common mtDNA template in CzechII mice
Abstract
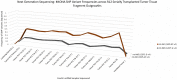
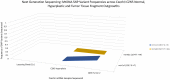
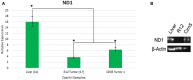
One major foundation of cancer etiology is the process of clonal expansion. The mechanisms underlying the complex process of a single cell leading to a clonal dominant tumor, are poorly understood. Our study aims to analyze mitochondrial DNA (mtDNA) for somatic single nucleotide polymorphisms (SNPs) variants, to determine if they are conserved throughout clonal expansion in mammary tissues and tumors. To test this hypothesis, we took advantage of a mouse mammary tumor virus (MMTV)-infected mouse model (CzechII). CzechII mouse mtDNA was extracted, from snap-frozen normal, hyperplastic, and tumor mammary epithelial outgrowth fragments. Next generation deep sequencing was used to determine if mtDNA "de novo" SNP variants are conserved during serial transplantation of both normal and neoplastic mammary clones. Our results support the conclusion that mtDNA "de novo" SNP variants are selected for and maintained during serial passaging of clonal phenotypically heterogeneous normal cellular populations; neoplastic cellular populations; metastatic clonal cellular populations and in individual tumor transplants, grown from the original metastatic tumor. In one case, a mammary tumor arising from a single cell, within a clonal hyperplastic outgrowth, contained only mtDNA copies, harboring a deleterious "de novo" SNP variant, suggesting that only one mtDNA template may act as a template for all mtDNA copies regardless of cell phenotype. This process has been attributed to "heteroplasmic-shifting". A process that is thought to result from selective pressure and may be responsible for pathogenic mutated mtDNA copies becoming homogeneous in clonal dominant oncogenic tissues.
Keywords: clonal expansion; mammary cancer; mitochondrial DNA; next-generation sequencing.
Conflict of interest statement
Conflicts of Interest The authors declare no potential conflicts of interest.
Figures




References
-
- Mashal RD, Lester SC, Sklar J. Clonal analysis by study of X chromosome inactivation in formalin-fixed paraffin embedded tissue. Cancer Res. 1993; 53:46776–46779. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

