Marked Changes in Gut Microbiota in Cardio-Surgical Intensive Care Patients: A Longitudinal Cohort Study
- PMID: 32010644
- PMCID: PMC6974539
- DOI: 10.3389/fcimb.2019.00467
Marked Changes in Gut Microbiota in Cardio-Surgical Intensive Care Patients: A Longitudinal Cohort Study
Abstract
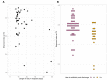
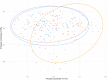
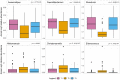
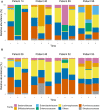
Background: Virtually no studies on the dynamics of the intestinal microbiota in patients admitted to the intensive care unit (ICU) are published, despite the increasingly recognized important role of microbiota on human physiology. Critical care patients undergo treatments that are known to influence the microbiota. However, dynamics and extent of such changes are not yet fully understood. To address this topic, we analyzed the microbiota before, during and after planned major cardio surgery that, for the first time, allowed us to follow the microbial dynamics of critical care patients. In this prospective, observational, longitudinal, single center study, we analyzed the fecal microbiota using 16S rRNA gene sequencing. Results: Samples of 97 patients admitted between April 2015 and November 2016 were included. In 32 patients, data of all three time points (before, during and after admission) were available for analysis. We found a large intra-individual variation in composition of gut microbiota. During admission, a significant change in microbial composition occurred in most patients, with a significant increase in pathobionts combined with a decrease in strictly anaerobic gut bacteria, typically beneficial for health. A lower bacterial diversity during admission was associated with longer hospitalization. In most patients analyzed at all three time points, the change in microbiota during hospital stay reverted to the original composition post-discharge. Conclusions: Our study shows that, even with a short ICU stay, patients present a significant change in microbial composition shortly after admission. The unique longitudinal setup of this study displayed a restoration of the microbiota in most patients to baseline composition post-discharge, which demonstrated its great restorative capacity. A relative decrease in benign or even beneficial bacteria and increase of pathobionts shifts the microbial balance in the gut, which could have clinical relevance. In future studies, the microbiota of ICU patients should be considered a good target for optimisation.
Keywords: 16S rRNA gene sequencing; critically ill; gut microbiota; intensive care unit; intestinal microbiota; longitudinal study.
Copyright © 2020 Aardema, Lisotto, Kurilshikov, Diepeveen, Friedrich, Sinha, de Smet and Harmsen.
Figures
References
-
- Bartram A. K., Lynch M. D., Stearns J. C., Moreno-Hagelsieb G., Neufeld J. D. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end illumina reads. Appl. Environ. Microbiol.. 77, 3846–3852. 10.1128/AEM.02772-10 - DOI - PMC - PubMed
-
- Buelow E., Bello González T. D., Fuentes S., de Steenhuijsen Piters W. A., Lahti L., Bayjanov J. R., et al. (2017). Comparative gut microbiota and resistome profiling of intensive care patients receiving selective digestive tract decontamination and healthy subjects. Microbiome 5:88. 10.1186/s40168-017-0309-z - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

