The Oral Microbiota May Have Influence on Oral Cancer
- PMID: 32010645
- PMCID: PMC6974454
- DOI: 10.3389/fcimb.2019.00476
The Oral Microbiota May Have Influence on Oral Cancer
Abstract
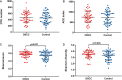
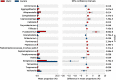
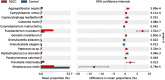
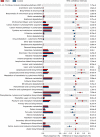
The oral microbiota plays an important role in the human microbiome and human health, and imbalances between microbes and their hosts can lead to oral and systemic diseases and chronic inflammation, which is usually caused by bacteria and contributes to cancer. There may be a relationship between oral bacteria and oral squamous cell carcinoma (OSCC); however, this relationship has not been thoroughly characterized. Therefore, in this study, we compared the microbiota compositions between tumor sites and opposite normal tissues in buccal mucosal of 50 patients with OSCC using the 16S rDNA sequencing. Richness and diversity of bacteria were significantly higher in tumor sites than in the control tissues. Cancer tissues were enriched in six families (Prevotellaceae, Fusobacteriaceae, Flavobacteriaceae, Lachnospiraceae, Peptostreptococcaceae, and Campylobacteraceae) and 13 genera, including Fusobacterium, Alloprevotella and Porphyromonas. At the species level, the abundances of Fusobacterium nucleatum, Prevotella intermedia, Aggregatibacter segnis, Capnocytophaga leadbetteri, Peptostreptococcus stomatis, and another five species were significantly increased, suggesting a potential association between these bacteria and OSCC. Furthermore, the functional prediction revealed that genes involved in bacterial chemotaxis, flagellar assembly and lipopolysaccharide (LPS) biosynthesis which are associated with various pathological processes, were significantly increased in the OSCC group. Overall, oral bacterial profiles showed significant difference between cancer sites and normal tissue of OSCC patients, which might be onsidered diagnostic markers and treatment targets. Our study has been registered in the Chinese clinical trial registry (ChiCTR1900025253, http://www.chictr.org.cn/index.aspx).
Keywords: 16S rDNA sequencing; Fusobacterium nucleatum; Peptostreptococcus stomatis; Prevotella intermedia; oral microbiota; oral squamous cell carcinoma.
Copyright © 2020 Zhang, Liu, Zheng and Zhang.
Figures

References
-
- Al-Hebshi N. N., Borgnakke W. S., Johnson N. W. (2019). The microbiome of oral squamous cell carcinomas: a functional perspective. Curr. Oral Health Rep. 6, 145–160. 10.1007/s40496-019-0215-5 - DOI
-
- Al-Hebshi N. N., Nasher A. T., Maryoud M. Y., Homeida H. E., Chen T., Idris A. M., et al. (2017). Inflammatory bacteriome featuring fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Sci. Rep. 7:1834. 10.1038/s41598-017-02079-3 - DOI - PMC - PubMed
-
- Binder Gallimidi A., Fischman S., Revach B., Bulvik R., Maliutina A., Rubinstein A. M., et al. (2015). Periodontal pathogens Porphyromonas gingivalis and fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 6, 22613–22623. 10.18632/oncotarget.4209 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

