Comparison of Three Xylose Pathways in Pseudomonas putida KT2440 for the Synthesis of Valuable Products
- PMID: 32010683
- PMCID: PMC6978631
- DOI: 10.3389/fbioe.2019.00480
Comparison of Three Xylose Pathways in Pseudomonas putida KT2440 for the Synthesis of Valuable Products
Abstract
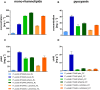
Pseudomonas putida KT2440 is a well-established chassis in industrial biotechnology. To increase the substrate spectrum, we implemented three alternative xylose utilization pathways, namely the Isomerase, Weimberg, and Dahms pathways. The synthetic operons contain genes from Escherichia coli and Pseudomonas taiwanensis. For isolating the Dahms pathway in P. putida KT2440 two genes (PP_2836 and PP_4283), encoding an endogenous enzyme of the Weimberg pathway and a regulator for glycolaldehyde degradation, were deleted. Before and after adaptive laboratory evolution, these strains were characterized in terms of growth and synthesis of mono-rhamnolipids and pyocyanin. The engineered strain using the Weimberg pathway reached the highest maximal growth rate of 0.30 h-1. After adaptive laboratory evolution the lag phase was reduced significantly. The highest titers of 720 mg L-1 mono-rhamnolipids and 30 mg L-1 pyocyanin were reached by the evolved strain using the Weimberg or an engineered strain using the Isomerase pathway, respectively. The different stoichiometries of the three xylose utilization pathways may allow engineering of tailored chassis for valuable bioproduct synthesis.
Keywords: Pseudomonas putida; flux balance analysis; heterologous production; metabolic engineering; phenazine; pyocyanin; rhamnolipid; xylose.
Copyright © 2020 Bator, Wittgens, Rosenau, Tiso and Blank.
Figures
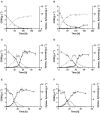
 , dark gray), formation of xylonate (
, dark gray), formation of xylonate ( , light gray), and cell dry weight (•, black) of (A)
P. putida KT2440, (B)
P. putida KT2440 pIso, (C)
P. putida KT2440 pWeim, (D)
P. putida KT2440ΔΔ pDahms, (E)
P. putida KT2440 pWeim2, and (F)
P. putida KT2440ΔΔ pDahms2. Error bars indicate deviation from the mean (n = 3).
, light gray), and cell dry weight (•, black) of (A)
P. putida KT2440, (B)
P. putida KT2440 pIso, (C)
P. putida KT2440 pWeim, (D)
P. putida KT2440ΔΔ pDahms, (E)
P. putida KT2440 pWeim2, and (F)
P. putida KT2440ΔΔ pDahms2. Error bars indicate deviation from the mean (n = 3).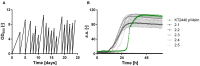


References
-
- Ashwell G., Wahba A. J., Hickman J. (1960). Uronic acid metabolism in bacteria. I. Purification and properties of uronic acid isomerase in Escherichia coli. J. Biol. Chem. 235, 1559–1565. - PubMed
-
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C. H., Bagdasarian M. M., Frey J., et al. (1981). Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16, 237–247. 10.1016/0378-1119(81)90080-9 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

