Interaction of yeast Rad51 and Rad52 relieves Rad52-mediated inhibition of de novo telomere addition
- PMID: 32012161
- PMCID: PMC7018233
- DOI: 10.1371/journal.pgen.1008608
Interaction of yeast Rad51 and Rad52 relieves Rad52-mediated inhibition of de novo telomere addition
Abstract
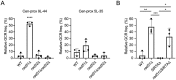
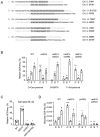
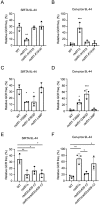
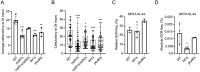
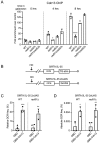
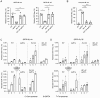
DNA double-strand breaks (DSBs) are toxic forms of DNA damage that must be repaired to maintain genome integrity. Telomerase can act upon a DSB to create a de novo telomere, a process that interferes with normal repair and creates terminal deletions. We previously identified sequences in Saccharomyces cerevisiae (SiRTAs; Sites of Repair-associated Telomere Addition) that undergo unusually high frequencies of de novo telomere addition, even when the original chromosome break is several kilobases distal to the eventual site of telomerase action. Association of the single-stranded telomere binding protein Cdc13 with a SiRTA is required to stimulate de novo telomere addition. Because extensive resection must occur prior to Cdc13 binding, we utilized these sites to monitor the effect of proteins involved in homologous recombination. We find that telomere addition is significantly reduced in the absence of the Rad51 recombinase, while loss of Rad52, required for Rad51 nucleoprotein filament formation, has no effect. Deletion of RAD52 suppresses the defect of the rad51Δ strain, suggesting that Rad52 inhibits de novo telomere addition in the absence of Rad51. The ability of Rad51 to counteract this effect of Rad52 does not require DNA binding by Rad51, but does require interaction between the two proteins, while the inhibitory effect of Rad52 depends on its interaction with Replication Protein A (RPA). Intriguingly, the genetic interactions we report between RAD51 and RAD52 are similar to those previously observed in the context of checkpoint adaptation. Forced recruitment of Cdc13 fully restores telomere addition in the absence of Rad51, suggesting that Rad52, through its interaction with RPA-coated single-stranded DNA, inhibits the ability of Cdc13 to bind and stimulate telomere addition. Loss of the Rad51-Rad52 interaction also stimulates a subset of Rad52-dependent microhomology-mediated repair (MHMR) events, consistent with the known ability of Rad51 to prevent single-strand annealing.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

