Staphylococcus epidermidis in feedings and feces of preterm neonates
- PMID: 32012172
- PMCID: PMC6996929
- DOI: 10.1371/journal.pone.0227823
Staphylococcus epidermidis in feedings and feces of preterm neonates
Abstract
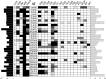
Staphylococcus epidermidis has emerged as the leading agent causing neonatal late-onset sepsis in preterm neonates; although the severity of the episodes caused by this species is often underestimated, it might exert relevant short- and long-term detrimental effects on neonatal outcomes. In this context, the objective of this study was to characterize a collection of S. epidermidis strains obtained from meconium and feces of preterm infants, and to assess the potential role of the enteral feeding tubes as potential reservoirs for this microorganism. A total of 26 preterm infants were enrolled in the study. Meconium and fecal samples were collected weekly during their first month of life (n = 92). Feeding samples were collected after their pass through the enteral feeding tubes (n = 84). S. epidermidis was present in the fecal samples of all the infants in, at least, one sampling time at concentrations ranging from 6.5 to 7.8 log10 CFU/g. Initially, 344 isolates were obtained and pulsed-field gel electrophoresis (PFGE) profiling allowed the reduction of the collection to 101 strains. Among them, multilocus sequence typing (MLST) profiling showed the presence of 32 different sequence types (ST). Globally, most of the STs to hospital-adapted high-risk clones and belonged to clonal complexes (CC) associated to the hospital environment, such as CC2. The virulence gene most commonly detected among the strains was altE. High resistance rates to macrolides and aminoglycosides were detected and 64% of the strains harboured the mecA gene, which was codified in SCCmec types. Our results indicates the existence of a complex and genetically diverse S. epidermidis population in the NICU environment. A better knowledge of S. epidermidis strains may help to devise strategies to avoid their conversion from symbiont to pathobiont microorganisms in the NICUs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Look Who's Talking: Host and Pathogen Drivers of Staphylococcus epidermidis Virulence in Neonatal Sepsis.Int J Mol Sci. 2022 Jan 13;23(2):860. doi: 10.3390/ijms23020860. Int J Mol Sci. 2022. PMID: 35055041 Free PMC article. Review.
-
Molecular Epidemiology of a Vancomycin-Intermediate Heteroresistant Staphylococcus epidermidis Outbreak in a Neonatal Intensive Care Unit.Antimicrob Agents Chemother. 2016 Sep 23;60(10):5673-81. doi: 10.1128/AAC.00726-16. Print 2016 Oct. Antimicrob Agents Chemother. 2016. PMID: 27401579 Free PMC article.
-
Serratia marcescens colonization in preterm neonates during their neonatal intensive care unit stay.Antimicrob Resist Infect Control. 2019 Aug 9;8:135. doi: 10.1186/s13756-019-0584-5. eCollection 2019. Antimicrob Resist Infect Control. 2019. PMID: 31413826 Free PMC article.
-
Molecular epidemiology of Staphylococcus epidermidis in neonatal intensive care units.APMIS. 2017 Jan;125(1):63-73. doi: 10.1111/apm.12637. Epub 2016 Nov 16. APMIS. 2017. PMID: 27859778 Clinical Trial.
-
Beyond sepsis: Staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity.Virulence. 2018 Jan 1;9(1):621-633. doi: 10.1080/21505594.2017.1419117. Virulence. 2018. PMID: 29405832 Free PMC article. Review.
Cited by
-
Altered Gut Microbiome and Fecal Immune Phenotype in Early Preterm Infants With Leaky Gut.Front Immunol. 2022 Feb 23;13:815046. doi: 10.3389/fimmu.2022.815046. eCollection 2022. Front Immunol. 2022. PMID: 35280991 Free PMC article.
-
Antibiotic susceptibility and phenotypic profile of Staphylococcus species isolated from different clinical samples from health facilities: A cross-sectional study.SAGE Open Med. 2024 Dec 17;12:20503121241306968. doi: 10.1177/20503121241306968. eCollection 2024. SAGE Open Med. 2024. PMID: 39698142 Free PMC article.
-
Profiling the response of individual gut microbes to free fatty acids (FFAs) found in human milk.J Funct Foods. 2025 Feb;125:106664. doi: 10.1016/j.jff.2025.106664. Epub 2025 Jan 8. J Funct Foods. 2025. PMID: 40051690 Free PMC article.
-
Look Who's Talking: Host and Pathogen Drivers of Staphylococcus epidermidis Virulence in Neonatal Sepsis.Int J Mol Sci. 2022 Jan 13;23(2):860. doi: 10.3390/ijms23020860. Int J Mol Sci. 2022. PMID: 35055041 Free PMC article. Review.
-
Antibiotic resistance genes in gut of breast-fed neonates born by caesarean section originate from breast milk and hospital ward air.BMC Microbiol. 2022 Jan 29;22(1):36. doi: 10.1186/s12866-022-02447-8. BMC Microbiol. 2022. PMID: 35093006 Free PMC article.
References
-
- Altuntas EG. Isolation, identification and characterization of Staphylococcus epidermidis in human milk. Food Sci Technol. 2015;60(1):36–41. 10.1016/j.lwt.2014.07.012 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

