Identification of genes of prognostic value in the ccRCC microenvironment from TCGA database
- PMID: 32012488
- PMCID: PMC7196483
- DOI: 10.1002/mgg3.1159
Identification of genes of prognostic value in the ccRCC microenvironment from TCGA database
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) is the most common pathological subtype of renal cell carcinoma. Bioinformatics analyses were used to screen candidate genes associated with the prognosis and microenvironment of ccRCC and elucidate the underlying molecular mechanisms of action.
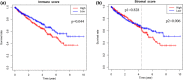
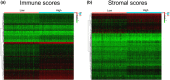
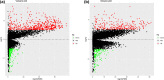
Methods: The gene expression profiles and clinical data of ccRCC patients were downloaded from The Cancer Genome Atlas database. The ESTIMATE algorithm was used to compute the immune and stromal scores of patients. Based on the median immune/stromal scores, all patients were sorted into low- and high-immune/stromal score groups. Differentially expressed genes (DEGs) were extracted from high- versus low-immune/stromal score groups and were described using functional annotations and protein-protein interaction (PPI) network.
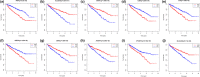
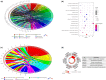
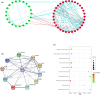
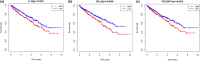
Results: Patients in the high-immune/stromal score group had poorer survival outcome. In total, 95 DEGs (48 upregulated and 47 downregulated genes) were screened from the gene expression profiles of patients with high immune and stromal scores. The genes were primarily involved in six signaling pathways. Among the 95 DEGs, 43 were markedly related to overall survival of patients. The PPI network identified the top 10 hub genes-CD19, CD79A, IL10, IGLL5, POU2AF1, CCL19, AMBP, CCL18, CCL21, and IGJ-and four modules. Enrichment analyses revealed that the genes in the most important module were involved in the B-cell receptor signaling pathway.
Conclusion: This study mainly revealed the relationship between the ccRCC microenvironment and prognosis of patients. These results also increase the understanding of how gene expression patterns can impact the prognosis and development of ccRCC by modulating the tumor microenvironment. The results could contribute to the search for ccRCC biomarkers and therapeutic targets.
Keywords: TCGA database; ccRCC; immune scores; microenvironment; stromal scores.
© 2020 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Data Mining of Prognostic Microenvironment-Related Genes in Clear Cell Renal Cell Carcinoma: A Study with TCGA Database.Dis Markers. 2019 Oct 30;2019:8901649. doi: 10.1155/2019/8901649. eCollection 2019. Dis Markers. 2019. PMID: 31781309 Free PMC article.
-
Comprehensive insights on pivotal prognostic signature involved in clear cell renal cell carcinoma microenvironment using the ESTIMATE algorithm.Cancer Med. 2020 Jun;9(12):4310-4323. doi: 10.1002/cam4.2983. Epub 2020 Apr 20. Cancer Med. 2020. PMID: 32311223 Free PMC article.
-
Deciphering Immune-Associated Genes to Predict Survival in Clear Cell Renal Cell Cancer.Biomed Res Int. 2019 Dec 7;2019:2506843. doi: 10.1155/2019/2506843. eCollection 2019. Biomed Res Int. 2019. PMID: 31886185 Free PMC article.
-
Clear cell renal cell carcinoma ontogeny and mechanisms of lethality.Nat Rev Nephrol. 2021 Apr;17(4):245-261. doi: 10.1038/s41581-020-00359-2. Epub 2020 Nov 3. Nat Rev Nephrol. 2021. PMID: 33144689 Free PMC article. Review.
-
The Tumor Immune Microenvironment in Clear Cell Renal Cell Carcinoma.Int J Mol Sci. 2023 Apr 27;24(9):7946. doi: 10.3390/ijms24097946. Int J Mol Sci. 2023. PMID: 37175653 Free PMC article. Review.
Cited by
-
Integrative Analysis of Immune-Related Genes in the Tumor Microenvironment of Renal Clear Cell Carcinoma and Renal Papillary Cell Carcinoma.Front Mol Biosci. 2021 Nov 23;8:760031. doi: 10.3389/fmolb.2021.760031. eCollection 2021. Front Mol Biosci. 2021. PMID: 34888353 Free PMC article.
-
Prognostic value of tumour microenvironment-related genes by TCGA database in rectal cancer.J Cell Mol Med. 2021 Jun;25(12):5811-5822. doi: 10.1111/jcmm.16547. Epub 2021 May 5. J Cell Mol Med. 2021. PMID: 33949771 Free PMC article.
-
Comprehensive Characterization of Immune Landscape Based on Tumor Microenvironment for Oral Squamous Cell Carcinoma Prognosis.Vaccines (Basel). 2022 Sep 14;10(9):1521. doi: 10.3390/vaccines10091521. Vaccines (Basel). 2022. PMID: 36146599 Free PMC article.
-
Hepcidin as a prognostic biomarker in clear cell renal cell carcinoma.Am J Cancer Res. 2022 Sep 15;12(9):4120-4139. eCollection 2022. Am J Cancer Res. 2022. PMID: 36225649 Free PMC article.
-
Increased expression of PSME2 is associated with clear cell renal cell carcinoma invasion by regulating BNIP3‑mediated autophagy.Int J Oncol. 2021 Dec;59(6):106. doi: 10.3892/ijo.2021.5286. Epub 2021 Nov 15. Int J Oncol. 2021. PMID: 34779489 Free PMC article.
References
-
- Agrawal, R. , Garg, A. , Benny Malgulwar, P. , Sharma, V. , Sarkar, C. , & Kulshreshtha, R. (2018). p53 and miR‐210 regulated NeuroD2, a neuronal basic helix‐loop‐helix transcription factor, is downregulated in glioblastoma patients and functions as a tumor suppressor under hypoxic microenvironment. International Journal of Cancer, 142(9), 1817–1828. 10.1002/ijc.31209 - DOI - PubMed
-
- Auer, R. L. , Starczynski, J. , McElwaine, S. , Bertoni, F. , Newland, A. C. , Fegan, C. D. , & Cotter, F. E. (2005). Identification of a potential role for POU2AF1 and BTG4 in the deletion of 11q23 in chronic lymphocytic leukemia. Genes, Chromosomes and Cancer, 43(1), 1–10. 10.1002/gcc.20159 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

