Exploring Protein Fold Space
- PMID: 32012781
- PMCID: PMC7072414
- DOI: 10.3390/biom10020193
Exploring Protein Fold Space
Abstract
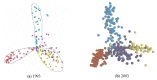
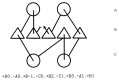
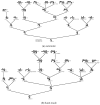
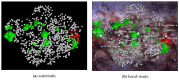
The model of protein folding proposed by Ptitsyn and colleagues involves the accretion of secondary structures around a nucleus. As developed by Efimov, this model also provides a useful way to view the relationships among structures. Although somewhat eclipsed by later databases based on the pairwise comparison of structures, Efimov's approach provides a guide for the more automatic comparison of proteins based on an encoding of their topology as a string. Being restricted to layers of secondary structures based on beta sheets, this too has limitations which are partly overcome by moving to a more generalised secondary structure lattice that can encompass both open and closed (barrel) sheets as well as helical packing of the type encoded by Murzin and Finkelstein on small polyhedra. Regular (crystalline) lattices, such as close-packed hexagonals, were found to be too limited so pseudo-latticses were investigated including those found in quasicrystals and the Bernal tetrahedron-based lattice that he used to represent liquid water. The Bernal lattice was considered best and used to generate model protein structures. These were much more numerous than those seen in Nature, posing the open question of why this might be.
Keywords: protein fold-space; protein structure comparison; secondary structure lattice.
Conflict of interest statement
The author declares no conflict of interest.
Figures







References
-
- Banner D.W., Bloomer A.C., Petsko G.A., Phillips D.C., Pogson C.I., Wilson I.A., Corran P.H., Furth A.J., Milman J.D., Offord R.E., et al. Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5Å resolution: Using amino acid sequence data. Nature. 1975;255:609–614. doi: 10.1038/255609a0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

