Characteristics of Human OAS1 Isoform Proteins
- PMID: 32013110
- PMCID: PMC7077331
- DOI: 10.3390/v12020152
Characteristics of Human OAS1 Isoform Proteins
Abstract
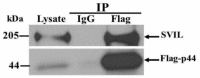
The human OAS1 (hOAS1) gene produces multiple possible isoforms due to alternative splicing events and sequence variation among individuals, some of which affect splicing. The unique C-terminal sequences of the hOAS1 isoforms could differentially affect synthetase activity, protein stability, protein partner interactions and/or cellular localization. Recombinant p41, p42, p44, p46, p48, p49 and p52 hOAS1 isoform proteins expressed in bacteria were each able to synthesize trimer and higher order 2'-5' linked oligoadenylates in vitro in response to poly(I:C). The p42, p44, p46, p48 and p52 isoform proteins were each able to induce RNase-mediated rRNA cleavage in response to poly(I:C) when overexpressed in HEK293 cells. The expressed levels of the p42 and p46 isoform proteins were higher than those of the other isoforms, suggesting increased stability in mammalian cells. In a yeast two-hybrid screen, Fibrillin1 (FBN1) was identified as a binding partner for hOAS1 p42 isoform, and Supervillin (SVIL) as a binding partner for the p44 isoform. The p44-SVIL interaction was supported by co-immunoprecipitation data from mammalian cells. The data suggest that the unique C-terminal regions of hOAS1 isoforms may mediate the recruitment of different partners, alternative functional capacities and/or different cellular localization.
Keywords: 2-5A; Fibrillin1; Supervillin; human OAS1 isoforms; rRNA cleavage.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Marie I., Hovanessian A.G. The 69-kDa 2-5A synthetase is composed of two homologous and adjacent functional domains. J. Biol. Chem. 1992;267:9933–9939. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

