Development of a Precision Medicine Workflow in Hematological Cancers, Aalborg University Hospital, Denmark
- PMID: 32013121
- PMCID: PMC7073219
- DOI: 10.3390/cancers12020312
Development of a Precision Medicine Workflow in Hematological Cancers, Aalborg University Hospital, Denmark
Abstract
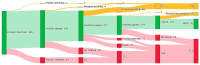
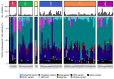
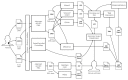
Within recent years, many precision cancer medicine initiatives have been developed. Most of these have focused on solid cancers, while the potential of precision medicine for patients with hematological malignancies, especially in the relapse situation, are less elucidated. Here, we present a demographic unbiased and observational prospective study at Aalborg University Hospital Denmark, referral site for 10% of the Danish population. We developed a hematological precision medicine workflow based on sequencing analysis of whole exome tumor DNA and RNA. All steps involved are outlined in detail, illustrating how the developed workflow can provide relevant molecular information to multidisciplinary teams. A group of 174 hematological patients with progressive disease or relapse was included in a non-interventional and population-based study, of which 92 patient samples were sequenced. Based on analysis of small nucleotide variants, copy number variants, and fusion transcripts, we found variants with potential and strong clinical relevance in 62% and 9.5% of the patients, respectively. The most frequently mutated genes in individual disease entities were in concordance with previous studies. We did not find tumor mutational burden or micro satellite instability to be informative in our hematologic patient cohort.
Keywords: bioinformatics workflow; hematology; next generation sequencing; precision medicine; somatic cancer variants; variant interpretation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Le Tourneau C., Delord J.P., Gonçalves A., Gavoille C., Dubot C., Isambert N., Campone M., Trédan O., Massiani M.A., Mauborgne C., et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

