Identification of QTL TGW12 responsible for grain weight in rice based on recombinant inbred line population crossed by wild rice (Oryza minuta) introgression line K1561 and indica rice G1025
- PMID: 32013862
- PMCID: PMC6998338
- DOI: 10.1186/s12863-020-0817-x
Identification of QTL TGW12 responsible for grain weight in rice based on recombinant inbred line population crossed by wild rice (Oryza minuta) introgression line K1561 and indica rice G1025
Abstract
Background: Limited genetic resource in the cultivated rice may hinder further yield improvement. Some valuable genes that contribute to rice yield may be lost or lacked in the cultivated rice. Identification of the quantitative trait locus (QTL) for yield-related traits such as thousand-grain weight (TGW) from wild rice speices is desired for rice yield improvement.
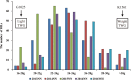
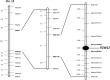
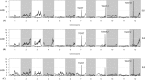
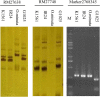
Results: In this study, sixteen TGW QTL were identified from a recombinant inbred line (RIL) population derived from the cross between the introgression line K1561 of Oryza minuta and the rice cultivar G1025. TGW12, One of most effective QTL was mapped to the region of 204.12 kb between the marker 2,768,345 and marker 2,853,491 of the specific locus amplified fragment (SLAF). The origin of TGW12 was tested using three markers nearby or within the TGW12 region, but not clarified yet. Our data indicated thirty-two open reading fragments (ORFs) were present in the region. RT-PCR analysis and sequence alignment showed that the coding domain sequences of ORF12, one MADS-box gene, in G1025 and K1561 were different due to alternative slicing, which caused premature transcription termination. The MADS-box gene was considered as a candidate of TGW12.
Conclusion: The effective QTL, TGW12, was mapped to a segment of 204.12 kb using RILs population and a MADS-box gene was identified among several candidate genes in the segment. The region of TGW12 should be further narrowed and creation of transgenic lines will reveal the gene function. TGW12 could be applied for improvement of TGW in breeding program.
Keywords: Introgression lines; SLAF; SSR; Transcription factor; Wild rice.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

