Atypical Salmonella enterica Serovars in Murine and Human Macrophage Infection Models
- PMID: 32014897
- PMCID: PMC7093118
- DOI: 10.1128/IAI.00353-19
Atypical Salmonella enterica Serovars in Murine and Human Macrophage Infection Models
Abstract
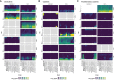
Nontyphoidal Salmonella species are globally disseminated pathogens and are the predominant cause of gastroenteritis. The pathogenesis of salmonellosis has been extensively studied using in vivo murine models and cell lines, typically challenged with Salmonella enterica serovar Typhimurium. Although S. enterica serovars Enteritidis and Typhimurium are responsible for most of the human infections reported to the Centers for Disease Control and Prevention (CDC), several other serovars also contribute to clinical cases of salmonellosis. Despite their epidemiological importance, little is known about their infection phenotypes. Here, we report the virulence characteristics and genomes of 10 atypical S. enterica serovars linked to multistate foodborne outbreaks in the United States. We show that the murine RAW 264.7 macrophage model of infection is unsuitable for inferring human-relevant differences in nontyphoidal Salmonella infections, whereas differentiated human THP-1 macrophages allowed these isolates to be further characterized in a more human-relevant context.
Keywords: Salmonella; Salmonella enterica; cytokines; infection; macrophages; pathogenicity; virulence; whole-genome sequencing.
Figures




References
-
- Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, Hall AJ, Keddy KH, Lake RJ, Lanata CF, Torgerson PR, Havelaar AH, Angulo FJ. 2015. World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis. PLoS Med 12:e1001921. doi: 10.1371/journal.pmed.1001921. - DOI - PMC - PubMed
-
- Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

