Retrograde trafficking of Argonaute 2 acts as a rate-limiting step for de novo miRNP formation on endoplasmic reticulum-attached polysomes in mammalian cells
- PMID: 32015087
- PMCID: PMC6998040
- DOI: 10.26508/lsa.201800161
Retrograde trafficking of Argonaute 2 acts as a rate-limiting step for de novo miRNP formation on endoplasmic reticulum-attached polysomes in mammalian cells
Abstract
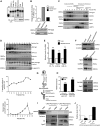
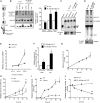
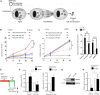
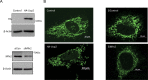
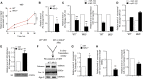
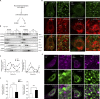
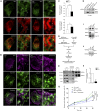
microRNAs are short regulatory RNAs in metazoan cells. Regulation of miRNA activity and abundance is evident in human cells where availability of target messages can influence miRNA biogenesis by augmenting the Dicer1-dependent processing of precursors to mature microRNAs. Requirement of subcellular compartmentalization of Ago2, the key component of miRNA repression machineries, for the controlled biogenesis of miRNPs is reported here. The process predominantly happens on the polysomes attached with the endoplasmic reticulum for which the subcellular Ago2 trafficking is found to be essential. Mitochondrial tethering of endoplasmic reticulum and its interaction with endosomes controls Ago2 availability. In cells with depolarized mitochondria, miRNA biogenesis gets impaired, which results in lowering of de novo-formed mature miRNA levels and accumulation of miRNA-free Ago2 on endosomes that fails to interact with Dicer1 and to traffic back to endoplasmic reticulum for de novo miRNA loading. Thus, mitochondria by sensing the cellular context regulates Ago2 trafficking at the subcellular level, which acts as a rate-limiting step in miRNA biogenesis process in mammalian cells.
© 2020 Bose et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
