A History of Cancer Research: Tumor Suppressor Genes
- PMID: 32015099
- PMCID: PMC6996451
- DOI: 10.1101/cshperspect.a035907
A History of Cancer Research: Tumor Suppressor Genes
Abstract
Tumor suppressor genes encode critical intracellular regulators, such as the retinoblastoma protein. They control processes including cell proliferation, cell survival, and responses to DNA damage and are frequently mutated in cancer. In this excerpt from his forthcoming book on the history of cancer research, Joe Lipsick looks back at the discovery of tumor suppressor genes, covering the early work on cell fusion by Henry Harris, Knudson's two-hit hypothesis, the genetic mapping studies that first identified the RB gene, and subsequent work on silencing.
Copyright © 2020 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures





References
-
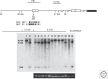
- Baylin S, Bestor TH. 2002. Altered methylation patterns in cancer cell genomes: Cause or consequence? Cancer Cell 1: 299–305. - PubMed
-
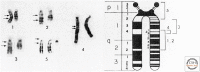
- Cavenee WK, Dryja TP, Phillips RA, Benedict WF, Godbout R, Gallie BL, Murphree AL, Strong LC, White RL. 1983. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305: 779–784. - PubMed
-
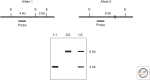
- Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. 1986. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 323: 643–646. - PubMed
-
- Harris H. 1993. How tumour suppressor genes were discovered. FASEB J 7: 978–979. - PubMed
-
- Knudson AG. 2001. Two genetic hits (more or less) to cancer. Nat Rev Cancer 1: 157–162. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
