Facilitative and synergistic interactions between fungal and plant viruses
- PMID: 32015104
- PMCID: PMC7035501
- DOI: 10.1073/pnas.1915996117
Facilitative and synergistic interactions between fungal and plant viruses
Abstract
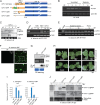
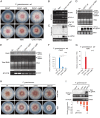
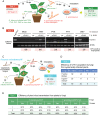
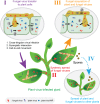
Plants and fungi are closely associated through parasitic or symbiotic relationships in which bidirectional exchanges of cellular contents occur. Recently, a plant virus was shown to be transmitted from a plant to a fungus, but it is unknown whether fungal viruses can also cross host barriers and spread to plants. In this study, we investigated the infectivity of Cryphonectria hypovirus 1 (CHV1, family Hypoviridae), a capsidless, positive-sense (+), single-stranded RNA (ssRNA) fungal virus in a model plant, Nicotiana tabacum CHV1 replicated in mechanically inoculated leaves but did not spread systemically, but coinoculation with an unrelated plant (+)ssRNA virus, tobacco mosaic virus (TMV, family Virgaviridae), or other plant RNA viruses, enabled CHV1 to systemically infect the plant. Likewise, CHV1 systemically infected transgenic plants expressing the TMV movement protein, and coinfection with TMV further enhanced CHV1 accumulation in these plants. Conversely, CHV1 infection increased TMV accumulation when TMV was introduced into a plant pathogenic fungus, Fusarium graminearum In the in planta F. graminearum inoculation experiment, we demonstrated that TMV infection of either the plant or the fungus enabled the horizontal transfer of CHV1 from the fungus to the plant, whereas CHV1 infection enhanced fungal acquisition of TMV. Our results demonstrate two-way facilitative interactions between the plant and fungal viruses that promote cross-kingdom virus infections and suggest the presence of plant-fungal-mediated routes for dissemination of fungal and plant viruses in nature.
Keywords: cross-kingdom; infection; mycovirus; plant virus.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Ghabrial S. A., Suzuki N., Viruses of plant pathogenic fungi. Annu. Rev. Phytopathol. 47, 353–384 (2009). - PubMed
-
- Ghabrial S. A., Castón J. R., Jiang D., Nibert M. L., Suzuki N., 50-plus years of fungal viruses. Virology 479–480, 356–368 (2015). - PubMed
-
- King A. M. Q., et al. , Changes to taxonomy and the international code of virus classification and nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 163, 2601–2631 (2018). - PubMed
-
- Hillman B. I., Annisa A., Suzuki N., “Viruses of plant-interacting fungi” in Advances in Virus Research, Kielian M., Mettenleiter T. C., M. J. R., Eds. (Elsevier, 2018), vol. 100, pp. 99–116. - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

