A conserved intratumoral regulatory T cell signature identifies 4-1BB as a pan-cancer target
- PMID: 32015231
- PMCID: PMC7269585
- DOI: 10.1172/JCI128672
A conserved intratumoral regulatory T cell signature identifies 4-1BB as a pan-cancer target
Abstract
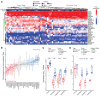
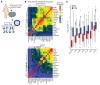
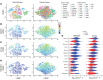
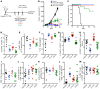
Despite advancements in targeting the immune checkpoints program cell death protein 1 (PD-1), programmed death ligand 1 (PD-L1), and cytotoxic T lymphocyte-associated protein 4 (CTLA-4) for cancer immunotherapy, a large number of patients and cancer types remain unresponsive. Current immunotherapies focus on modulating an antitumor immune response by directly or indirectly expanding antitumor CD8 T cells. A complementary strategy might involve inhibition of Tregs that otherwise suppress antitumor immune responses. Here, we sought to identify functional immune molecules preferentially expressed on tumor-infiltrating Tregs. Using genome-wide RNA-Seq analysis of purified Tregs sorted from multiple human cancer types, we identified a conserved Treg immune checkpoint signature. Using immunocompetent murine tumor models, we found that antibody-mediated depletion of 4-1BB-expressing cells (4-1BB is also known as TNFRSF9 or CD137) decreased tumor growth without negatively affecting CD8 T cell function. Furthermore, we found that the immune checkpoint 4-1BB had a high selectivity for human tumor Tregs and was associated with worse survival outcomes in patients with multiple tumor types. Thus, antibody-mediated depletion of 4-1BB-expressing Tregs represents a strategy with potential activity across cancer types.
Keywords: Cancer immunotherapy; Immunology; Immunotherapy; Oncology; T cells.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

