Two-stage electro-mechanical coupling of a KV channel in voltage-dependent activation
- PMID: 32015334
- PMCID: PMC6997178
- DOI: 10.1038/s41467-020-14406-w
Two-stage electro-mechanical coupling of a KV channel in voltage-dependent activation
Abstract
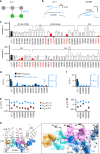
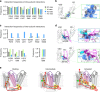
In voltage-gated potassium (KV) channels, the voltage-sensing domain (VSD) undergoes sequential activation from the resting state to the intermediate state and activated state to trigger pore opening via electro-mechanical (E-M) coupling. However, the spatial and temporal details underlying E-M coupling remain elusive. Here, utilizing KV7.1's unique two open states, we report a two-stage E-M coupling mechanism in voltage-dependent gating of KV7.1 as triggered by VSD activations to the intermediate and then activated state. When the S4 segment transitions to the intermediate state, the hand-like C-terminus of the VSD-pore linker (S4-S5L) interacts with the pore in the same subunit. When S4 then proceeds to the fully-activated state, the elbow-like hinge between S4 and S4-S5L engages with the pore of the neighboring subunit to activate conductance. This two-stage hand-and-elbow gating mechanism elucidates distinct tissue-specific modulations, pharmacology, and disease pathogenesis of KV7.1, and likely applies to numerous domain-swapped KV channels.
Conflict of interest statement
J.S. and J.C. are cofounders of a startup company VivoCor LLC, which is targeting
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

