RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root
- PMID: 32015352
- PMCID: PMC6997459
- DOI: 10.1038/s41598-020-57884-0
RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root
Erratum in
-
Author Correction: RNA-Seq reveals novel genes and pathways associated with hypoxia duration and tolerance in tomato root.Sci Rep. 2020 Nov 20;10(1):20642. doi: 10.1038/s41598-020-75106-5. Sci Rep. 2020. PMID: 33219234 Free PMC article.
Abstract
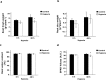
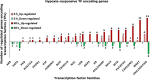
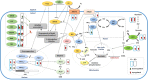
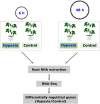
Due to climate change, economically important crop plants will encounter flooding periods causing hypoxic stress more frequently. This may lead to reduced yields and endanger food security. As roots are the first organ to be affected by hypoxia, the ability to sense and respond to hypoxic stress is crucial. At the molecular level, therefore, fine-tuning the regulation of gene expression in the root is essential for hypoxia tolerance. Using an RNA-Seq approach, we investigated transcriptome modulation in tomato roots of the cultivar 'Moneymaker', in response to short- (6 h) and long-term (48 h) hypoxia. Hypoxia duration appeared to have a significant impact on gene expression such that the roots of five weeks old tomato plants showed a distinct time-dependent transcriptome response. We observed expression changes in 267 and 1421 genes under short- and long-term hypoxia, respectively. Among these, 243 genes experienced changed expression at both time points. We identified tomato genes with a potential role in aerenchyma formation which facilitates oxygen transport and may act as an escape mechanism enabling hypoxia tolerance. Moreover, we identified differentially regulated genes related to carbon and amino acid metabolism and redox homeostasis. Of particular interest were the differentially regulated transcription factors, which act as master regulators of downstream target genes involved in responses to short and/or long-term hypoxia. Our data suggest a temporal metabolic and anatomic adjustment to hypoxia in tomato root which requires further investigation. We propose that the regulated genes identified in this study are good candidates for further studies regarding hypoxia tolerance in tomato or other crops.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

