Cell-mediated immune response and protective efficacy of porcine reproductive and respiratory syndrome virus modified-live vaccines against co-challenge with PRRSV-1 and PRRSV-2
- PMID: 32015495
- PMCID: PMC6997162
- DOI: 10.1038/s41598-020-58626-y
Cell-mediated immune response and protective efficacy of porcine reproductive and respiratory syndrome virus modified-live vaccines against co-challenge with PRRSV-1 and PRRSV-2
Abstract
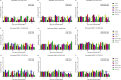
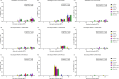
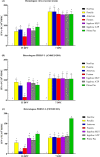
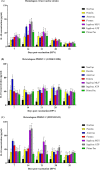
Cell-mediated immunity (CMI), IL-10, and the protective efficacy of modified-live porcine reproductive and respiratory syndrome virus (PRRSV) vaccines (MLV) against co-challenge with PRRSV-1 and PRRSV-2 (HP-PRRSV) were investigated. Seventy, PRRSV-free, 3-week old, pigs were allocated into 7 groups. Six groups were intramuscularly vaccinated with MLV, including Porcilis (PRRSV-1 MLV, MSD Animal Health, The Netherlands), Amervac (PRRSV-1 MLV, Laboratorios Hipra, Spain), Fostera (PRRSV-2 MLV, Zoetis, USA), Ingelvac PRRS MLV and Ingelvac PRRS ATP (PRRSV-2, Boehringer Ingelheim, USA), and Prime Pac PRRS (PRRSV-2 MLV, MSD Animal Health, The Netherlands). Unvaccinated pigs were left as control. Lymphocyte proliferative response, IL-10 and IFN-γ production were determined. At 35 days post-vaccination (DPV), all pigs were inoculated intranasally with 2 ml of each PRRSV-1 (105.4 TCID50/ml) and PRRSV-2 (105.2 TCID50/ml, HP-PRRSV). Following challenge, sera were quantitatively assayed for PRRSV RNA. Pigs were necropsied at 7 days post-challenge. Viremia, macro- and microscopic lung lesion together with PRRSV antigen presence were evaluated in lung tissues. The results demonstrated that, regardless of vaccine genotype, CMI induced by all MLVs was relatively slow. Increased production of IL-10 in all vaccinated groups was observed at 7 and 14 DPV. Pigs in Amervac, Ingelvac MLV and Ingelvac ATP groups had significantly higher levels of IL-10 compared to Porcilis, Fostera and Prime Pac groups at 7 and 14 DPV. Following challenge, regardless to vaccine genotype, vaccinated pigs had significantly lower lung lesion scores and PRRSV antigens than those in the control group. Both PRRSV-1 and PRRSV-2 RNA were significantly reduced. Prime Pac pigs had lowest PRRSV-1 and PRRSV-2 RNA in serum, and micro- and macroscopic lung lesion scores (p < 0.05) compared to other vaccinated groups. In conclusion, PRRSV MLVs, regardless of vaccine genotype, can reduce viremia and lung lesions following co-challenge with PRRSV-1 and PRRSV-2 (HP-PRRSV). The main difference between PRRSV MLV is the production of IL-10 following vaccination.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997;142:629–633. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

