Keth-seq for transcriptome-wide RNA structure mapping
- PMID: 32015521
- PMCID: PMC7182492
- DOI: 10.1038/s41589-019-0459-3
Keth-seq for transcriptome-wide RNA structure mapping
Abstract
RNA secondary structure is critical to RNA regulation and function. We report a new N3-kethoxal reagent that allows fast and reversible labeling of single-stranded guanine bases in live cells. This N3-kethoxal-based chemistry allows efficient RNA labeling under mild conditions and transcriptome-wide RNA secondary structure mapping.
Conflict of interest statement
Competing financial interests:
C.H. is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc., and a shareholder of Epican Genetech.
Figures

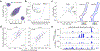
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

