A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder
- PMID: 32015540
- PMCID: PMC7065955
- DOI: 10.1038/s41593-019-0578-x
A myelin-related transcriptomic profile is shared by Pitt-Hopkins syndrome models and human autism spectrum disorder
Abstract
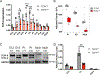
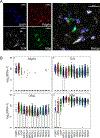
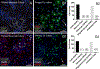
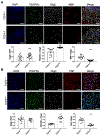
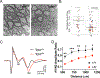
Autism spectrum disorder (ASD) is genetically heterogeneous with convergent symptomatology, suggesting common dysregulated pathways. In this study, we analyzed brain transcriptional changes in five mouse models of Pitt-Hopkins syndrome (PTHS), a syndromic form of ASD caused by mutations in the TCF4 gene, but not the TCF7L2 gene. Analyses of differentially expressed genes (DEGs) highlighted oligodendrocyte (OL) dysregulation, which we confirmed in two additional mouse models of syndromic ASD (Ptenm3m4/m3m4 and Mecp2tm1.1Bird). The PTHS mouse models showed cell-autonomous reductions in OL numbers and myelination, functionally confirming OL transcriptional signatures. We also integrated PTHS mouse model DEGs with human idiopathic ASD postmortem brain RNA-sequencing data and found significant enrichment of overlapping DEGs and common myelination-associated pathways. Notably, DEGs from syndromic ASD mouse models and reduced deconvoluted OL numbers distinguished human idiopathic ASD cases from controls across three postmortem brain data sets. These results implicate disruptions in OL biology as a cellular mechanism in ASD pathology.
Conflict of interest statement
Competing Interest Statement
The authors declare that they have no competing financial interests.
Figures



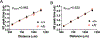









References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

