Identification of a potentially functional circRNA-miRNA-mRNA regulatory network for investigating pathogenesis and providing possible biomarkers of bladder cancer
- PMID: 32015691
- PMCID: PMC6990554
- DOI: 10.1186/s12935-020-1108-3
Identification of a potentially functional circRNA-miRNA-mRNA regulatory network for investigating pathogenesis and providing possible biomarkers of bladder cancer
Abstract
Background: Circular RNAs (circRNAs) have received considerable attention in human cancer research. However, many circRNAs remain to be detected. In our study, we determined novel circRNAs and investigated their effects on bladder cancer (BCa).
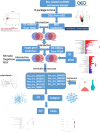
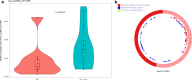
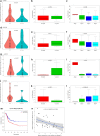
Methods: Microarray dataset GSE92675 was downloaded from Gene Expression Omnibus (GEO). Then, we combined computational biology with quantitative real-time polymerase chain reaction (qRT-PCR) to select related circRNAs in BCa. The selected circRNA-microRNA (miRNA)-messenger RNA (mRNA) regulatory subnetwork was determined by Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
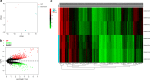
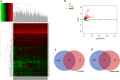
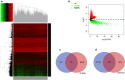
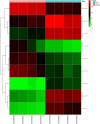
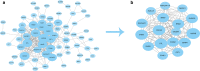
Results: The regulatory network constructed from the microarray dataset (GSE92675) contained 49 differentially expressed circRNAs (DECs). GO and KEGG analyses showed that the MAPK and PI3K-AKT signaling pathways were statistically significant. On the basis of qRT-PCR and the degree value calculated by the cytoHubba plugin of Cytoscape, hsa_circ_0011385 was finally confirmed. The subnetwork around hsa_circ_0011385 was constructed. In addition, we created a protein-protein interaction (PPI) network composed of 67 nodes and 274 edges after removing independent nodes. GO and KEGG analyses showed that hubgenes were involved in cell cycle activities. Moreover, they could be regulated by miRNAs and play an eventful role in BCa pathogenesis.
Conclusions: We proposed a novel circRNA-miRNA-mRNA network related to BCa pathogenesis. This network might be a new molecular biomarker and could be used to develop potential treatment strategies for BCa.
Keywords: Biomarker; Bladder cancer; ceRNA; circRNA.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures






Similar articles
-
Construction of Circular RNA-MicroRNA-Messenger RNA Regulatory Network of Recurrent Implantation Failure to Explore Its Potential Pathogenesis.Front Genet. 2021 Feb 16;11:627459. doi: 10.3389/fgene.2020.627459. eCollection 2020. Front Genet. 2021. PMID: 33664765 Free PMC article.
-
Construction of a circRNA-miRNA-mRNA network based on differentially co-expressed circular RNA in gastric cancer tissue and plasma by bioinformatics analysis.World J Surg Oncol. 2022 Feb 14;20(1):34. doi: 10.1186/s12957-022-02503-7. World J Surg Oncol. 2022. PMID: 35164778 Free PMC article.
-
Identification of Potentially Functional CircRNA-miRNA-mRNA Regulatory Network in Hepatocellular Carcinoma by Integrated Microarray Analysis.Med Sci Monit Basic Res. 2018 Apr 30;24:70-78. doi: 10.12659/MSMBR.909737. Med Sci Monit Basic Res. 2018. PMID: 29706616 Free PMC article.
-
Identification of the circRNA-miRNA-mRNA regulatory network and its prognostic effect in colorectal cancer.World J Clin Cases. 2021 Jun 26;9(18):4520-4541. doi: 10.12998/wjcc.v9.i18.4520. World J Clin Cases. 2021. PMID: 34222420 Free PMC article.
-
Identification of Serum Exosome-Derived circRNA-miRNA-TF-mRNA Regulatory Network in Postmenopausal Osteoporosis Using Bioinformatics Analysis and Validation in Peripheral Blood-Derived Mononuclear Cells.Front Endocrinol (Lausanne). 2022 Jun 9;13:899503. doi: 10.3389/fendo.2022.899503. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35757392 Free PMC article.
Cited by
-
A network pharmacology approach to determine the underlying mechanisms of action of Yishen Tongluo formula for the treatment of oligoasthenozoospermia.PLoS One. 2021 Jun 21;16(6):e0252906. doi: 10.1371/journal.pone.0252906. eCollection 2021. PLoS One. 2021. PMID: 34153045 Free PMC article.
-
Circular RNA regulates the onset and progression of cancer through the mitogen-activated protein kinase signaling pathway.Oncol Lett. 2021 Dec;22(6):817. doi: 10.3892/ol.2021.13078. Epub 2021 Oct 5. Oncol Lett. 2021. PMID: 34671431 Free PMC article. Review.
-
Identification of potential circRNAs and circRNA-miRNA-mRNA regulatory network in the development of diabetic foot ulcers by integrated bioinformatics analysis.Int Wound J. 2021 Jun;18(3):323-331. doi: 10.1111/iwj.13535. Epub 2020 Dec 13. Int Wound J. 2021. PMID: 33314661 Free PMC article.
-
Construction of Circular RNA-MicroRNA-Messenger RNA Regulatory Network of Recurrent Implantation Failure to Explore Its Potential Pathogenesis.Front Genet. 2021 Feb 16;11:627459. doi: 10.3389/fgene.2020.627459. eCollection 2020. Front Genet. 2021. PMID: 33664765 Free PMC article.
-
Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer.Mol Cancer. 2021 Jan 4;20(1):4. doi: 10.1186/s12943-020-01300-8. Mol Cancer. 2021. PMID: 33397425 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

