Small RNA Sequencing Reveals Exosomal miRNAs Involved in the Treatment of Asthma by Scorpio and Centipede
- PMID: 32016112
- PMCID: PMC6985928
- DOI: 10.1155/2020/1061407
Small RNA Sequencing Reveals Exosomal miRNAs Involved in the Treatment of Asthma by Scorpio and Centipede
Abstract
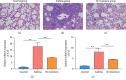
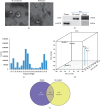
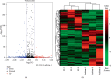
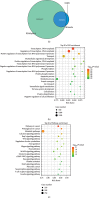
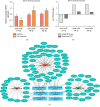
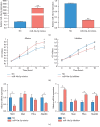
Asthma is a common respiratory disease with inflammation in the lungs. Exosomes and microRNAs (miRNAs) play crucial role in inflammation, whereas the role of exosomal miRNA in asthma remains unknown. Here, we aimed to identify the key exosomal miRNAs and their underlying mechanisms involved in scorpio and centipede (SC) treatment in asthma. Eighteen mice were randomly divided into three groups: control group, asthma group, and SC treatment group. Effect of SC was assessed by hematoxylin-eosin staining and real-time PCR. Exosomes from asthma and SC treatment groups were analyzed by small RNA-seq. Results revealed SC significantly alleviated the pathogenesis of asthma and suppressed the release of inflammatory cytokines. A total of 328 exosomal miRNAs were differentially expressed between the exosomes from asthma and SC-treated mice, including 118 up- and 210 downregulated in SC-treated mice. The altered exosomal miRNAs were primarily involved in the function of transcription, apoptotic process, and cell adhesion; and pathway of calcium, Wnt, and MAPK signaling. Real-time PCR verified exosomal miR-147 was downregulated, while miR-98-5p and miR-10a-5p were upregulated in SC-treated mice compared to asthma mice. Moreover, the target genes of miR-147-3p, miR-98-5p, and miR-10a-5p were mainly enriched in Wnt and MAPK inflammatory signaling. miR-10a-5p promoted the proliferation of mouse lung epithelial cells and downregulated the expression of Nfat5 and Map2k6. These data suggest SC-induced exosomal miRNAs might mediate the inflammatory signaling and might be involved in the SC treatment in asthma. The exosomal miRNAs might be promising candidates for the treatment of asthma.
Copyright © 2020 Binqing Tang et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
Similar articles
-
Scorpion and centipede alleviates severe asthma through M2 macrophage-derived exosomal miR-30b-5p.Aging (Albany NY). 2022 May 2;14(9):3921-3940. doi: 10.18632/aging.204053. Epub 2022 May 2. Aging (Albany NY). 2022. PMID: 35500231 Free PMC article.
-
Exploring the pathogenesis of type 2 diabetes mellitus intestinal damp-heat syndrome and the therapeutic effect of Gegen Qinlian Decoction from the perspective of exosomal miRNA.J Ethnopharmacol. 2022 Mar 1;285:114786. doi: 10.1016/j.jep.2021.114786. Epub 2021 Nov 8. J Ethnopharmacol. 2022. PMID: 34763043
-
Exosomal miR-1228-5p down-regulates DUSP22 to promotes cell proliferation and migration in small cell lung cancer.Life Sci. 2024 Aug 15;351:122787. doi: 10.1016/j.lfs.2024.122787. Epub 2024 Jun 6. Life Sci. 2024. PMID: 38851418
-
MicroRNAs: future biomarkers and targets of therapy in asthma?Curr Opin Pulm Med. 2020 May;26(3):285-292. doi: 10.1097/MCP.0000000000000673. Curr Opin Pulm Med. 2020. PMID: 32101904 Review.
-
MicroRNAs: Potential Biomarkers and Targets of Therapy in Allergic Diseases?Arch Immunol Ther Exp (Warsz). 2019 Aug;67(4):213-223. doi: 10.1007/s00005-019-00547-4. Epub 2019 May 28. Arch Immunol Ther Exp (Warsz). 2019. PMID: 31139837 Free PMC article. Review.
Cited by
-
Promising Therapeutic Functions of Bone Marrow Mesenchymal Stem Cells Derived-Exosome in Asthma.Can Respir J. 2022 Dec 20;2022:1485719. doi: 10.1155/2022/1485719. eCollection 2022. Can Respir J. 2022. PMID: 36582191 Free PMC article. Review.
-
Scorpion and centipede alleviates severe asthma through M2 macrophage-derived exosomal miR-30b-5p.Aging (Albany NY). 2022 May 2;14(9):3921-3940. doi: 10.18632/aging.204053. Epub 2022 May 2. Aging (Albany NY). 2022. PMID: 35500231 Free PMC article.
-
Emu-miR-10a-5p in Echinococcus multilocularis-derived-extracellular vesicles alleviates airway inflammation in mice with allergic asthma by inhibiting macrophage M2a polarization through LIF-mediated JAK1-STAT3 signaling.Front Immunol. 2025 May 27;16:1577349. doi: 10.3389/fimmu.2025.1577349. eCollection 2025. Front Immunol. 2025. PMID: 40496853 Free PMC article.
-
Non-Exosomal and Exosome-Derived miRNAs as Promising Biomarkers in Canine Mammary Cancer.Life (Basel). 2022 Apr 1;12(4):524. doi: 10.3390/life12040524. Life (Basel). 2022. PMID: 35455015 Free PMC article. Review.
-
Extracellular Vesicles and Asthma-More Than Just a Co-Existence.Int J Mol Sci. 2021 May 7;22(9):4984. doi: 10.3390/ijms22094984. Int J Mol Sci. 2021. PMID: 34067156 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

