Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway
- PMID: 32016387
- PMCID: PMC7492770
- DOI: 10.1093/jnci/djaa017
Radiomics Response Signature for Identification of Metastatic Colorectal Cancer Sensitive to Therapies Targeting EGFR Pathway
Abstract
Background: The authors sought to forecast survival and enhance treatment decisions for patients with liver metastatic colorectal cancer by using on-treatment radiomics signature to predict tumor sensitiveness to irinotecan, 5-fluorouracil, and leucovorin (FOLFIRI) alone (F) or in combination with cetuximab (FC).
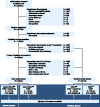
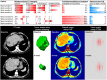
Methods: We retrospectively analyzed 667 metastatic colorectal cancer patients treated with F or FC. Computed tomography quality was classified as high (HQ) or standard (SD). Four datasets were created using the nomenclature (treatment) - (quality). Patients were randomly assigned (2:1) to training or validation sets: FCHQ: 78:38, FCSD: 124:62, FHQ: 78:51, FSD: 158:78. Four tumor-imaging biomarkers measured quantitative radiomics changes between standard of care computed tomography scans at baseline and 8 weeks. Using machine learning, the performance of the signature to classify tumors as treatment sensitive or treatment insensitive was trained and validated using receiver operating characteristic (ROC) curves. Hazard ratio and Cox regression models evaluated association with overall survival (OS).
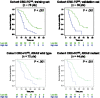
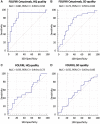
Results: The signature (area under the ROC curve [95% confidence interval (CI)]) used temporal decrease in tumor spatial heterogeneity plus boundary infiltration to successfully predict sensitivity to antiepidermal growth factor receptor therapy (FCHQ: 0.80 [95% CI = 0.69 to 0.94], FCSD: 0.72 [95% CI = 0.59 to 0.83]) but failed with chemotherapy (FHQ: 0.59 [95% CI = 0.44 to 0.72], FSD: 0.55 [95% CI = 0.43 to 0.66]). In cetuximab-containing sets, radiomics signature outperformed existing biomarkers (KRAS-mutational status, and tumor shrinkage by RECIST 1.1) for detection of treatment sensitivity and was strongly associated with OS (two-sided P < .005).
Conclusions: Radiomics response signature can serve as an intermediate surrogate marker of OS. The signature outperformed known biomarkers in providing an early prediction of treatment sensitivity and could be used to guide cetuximab treatment continuation decisions.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures

Comment in
-
The Evolving Status of Radiomics.J Natl Cancer Inst. 2020 Sep 1;112(9):869-870. doi: 10.1093/jnci/djaa018. J Natl Cancer Inst. 2020. PMID: 32016420 Free PMC article. No abstract available.
References
-
- Van Cutsem E, Cervantes A, Nordlinger B,. et al. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(suppl 3):iii1–9. - PubMed
-
- Schwartz LH, Mazumdar M, Brown W, et al. Variability in response assessment in solid tumors: effect of number of lesions chosen for measurement. Clin Cancer Res. 2003;9(12):4318–4323. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

