Structure-Guided Design and In-Cell Target Profiling of a Cell-Active Target Engagement Probe for PARP Inhibitors
- PMID: 32017532
- PMCID: PMC7146755
- DOI: 10.1021/acschembio.9b00963
Structure-Guided Design and In-Cell Target Profiling of a Cell-Active Target Engagement Probe for PARP Inhibitors
Abstract
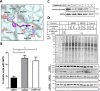
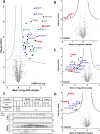
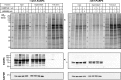
Inhibition of the poly(ADP-ribose) polymerase (PARP) family of enzymes has become an attractive therapeutic strategy in oncology and beyond; however, chemical tools to profile PARP engagement in live cells are lacking. Herein, we report the design and application of PARPYnD, the first photoaffinity probe (AfBP) for PARP enzymes based on triple PARP1/2/6 inhibitor AZ9482, which induces multipolar spindle (MPS) formation in breast cancer cells. PARPYnD is a robust tool for profiling PARP1/2 and is used to profile clinical PARP inhibitor olaparib, identifying several novel off-target proteins. Surprisingly, while PARPYnD can enrich recombinant PARP6 spiked into cellular lysates and inhibits PARP6 in cell-free assays, it does not label PARP6 in intact cells. These data highlight an intriguing biomolecular disparity between recombinant and endogenous PARP6. PARPYnD provides a new approach to expand our knowledge of the targets of this class of compounds and the mechanisms of action of PARP inhibitors in cancer.
Conflict of interest statement
The authors declare the following competing financial interest(s): This work was partially funded by AstraZeneca, and P.H., P.P., J.S.S., and J.W.J. are all employees of AstraZeneca. E.W.T. is a Director and shareholder of Myricx Pharma Ltd.
Figures

References
-
- Yang C.-S.; Jividen K.; Spencer A.; Dworak N.; Ni L.; Oostdyk L. T.; Chatterjee M.; Kuśmider B.; Reon B.; Parlak M.; Gorbunova V.; Abbas T.; Jeffery E.; Sherman N. E.; Paschal B. M. (2017) Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516. 10.1016/j.molcel.2017.04.028. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

