Molecular analysis of lymphoid tissue from rhesus macaque rhadinovirus-infected monkeys identifies alterations in host genes associated with oncogenesis
- PMID: 32017809
- PMCID: PMC6999886
- DOI: 10.1371/journal.pone.0228484
Molecular analysis of lymphoid tissue from rhesus macaque rhadinovirus-infected monkeys identifies alterations in host genes associated with oncogenesis
Abstract
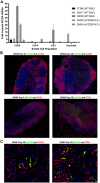
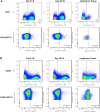
Rhesus macaque (RM) rhadinovirus (RRV) is a simian gamma-2 herpesvirus closely related to human Kaposi's sarcoma-associated herpesvirus (KSHV). RRV is associated with the development of diseases in simian immunodeficiency virus (SIV) co-infected RM that resemble KSHV-associated pathologies observed in HIV-infected humans, including B cell lymphoproliferative disorders (LPD) and lymphoma. Importantly, how de novo KSHV infection affects the expression of host genes in humans, and how these alterations in gene expression affect viral replication, latency, and disease is unknown. The utility of the RRV/RM infection model provides a novel approach to address these questions in vivo, and utilizing the RRV bacterial artificial chromosome (BAC) system, the effects of specific viral genes on host gene expression patterns can also be explored. To gain insight into the effects of RRV infection on global host gene expression patterns in vivo, and to simultaneously assess the contributions of the immune inhibitory viral CD200 (vCD200) molecule to host gene regulation, RNA-seq was performed on pre- and post-infection lymph node (LN) biopsy samples from RM infected with either BAC-derived WT (n = 4) or vCD200 mutant RRV (n = 4). A variety of genes were identified as being altered in LN tissue samples due to RRV infection, including cancer-associated genes activation-induced cytidine deaminase (AICDA), glypican-1 (GPC1), CX3C chemokine receptor 1 (CX3CR1), and Ras dexamethasone-induced 1 (RasD1). Further analyses also indicate that GPC1 may be associated with lymphomagenesis. Finally, comparison of infection groups identified the differential expression of host gene thioredoxin interacting protein (TXNIP), suggesting a possible mechanism by which vCD200 negatively affects RRV viral loads in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Wong SW, Bergquam EP, Swanson RM, Lee FW, Shiigi SM, Avery NA, et al. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi’s sarcoma-associated herpesvirus. J Exp Med. 1999;190(6):827–40. 10.1084/jem.190.6.827 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

